Intron-skipping siRNAs¶
Evidence to support the hypothesis that RNAi is triggered against spliced transcripts, rather than by introns as proposed by [Dumesic-2013], is provided by reads that cross splice sites after removal of the intron-sequence in line with mRNA-Seq data.
Counting of intron-like gaps has been based on the presence of N within the cigar string for each alignment. In the tutorial on ‘Cryptococcus siRNAs’ it is described how some of these intron-skipping reads stand out for mutant strains rde1Δ and rde2Δ. Gaps of ~60 nt are predicted for alignments in the absence of Rde1 as shown in Figure 12 (lanes re1; in the diagram showing mapped reads bridging a gap; middle row, left panel), while in the absence of Rde2 multimapping reads over spliced introns of ~73 nt were enriched (lanes re2 in the same Figure, right panels of the middle or bottom row). These reads can be retrieved from the bam files directly with Samtools. The IDs for the re1 samples are SRR8697586 and SRR8697587, and for re2 SRR8697588 and SRR8697589. We will check the alignments for reads that were mapped across the putative introns of 68 and 73 nt.
But first, to directly support the argument that transcripts have been spliced prior to the onset of an RNAi response, we can identify reads across spliced introns in CNAG_03231 (former CNAG_07721) in the wild type, SRR646636, analyzed as a ‘model’ siRNA-target by [Dumesic-2013].
Introns in CNAG_03231¶
Figure 1 displays the CNAG_03231 locus in the IGB genome browser (after loading bedgraphs for wild type and ago1Δ RNA-Seq data [Wallace-2020] and for siRNAs [Dumesic-2013]).
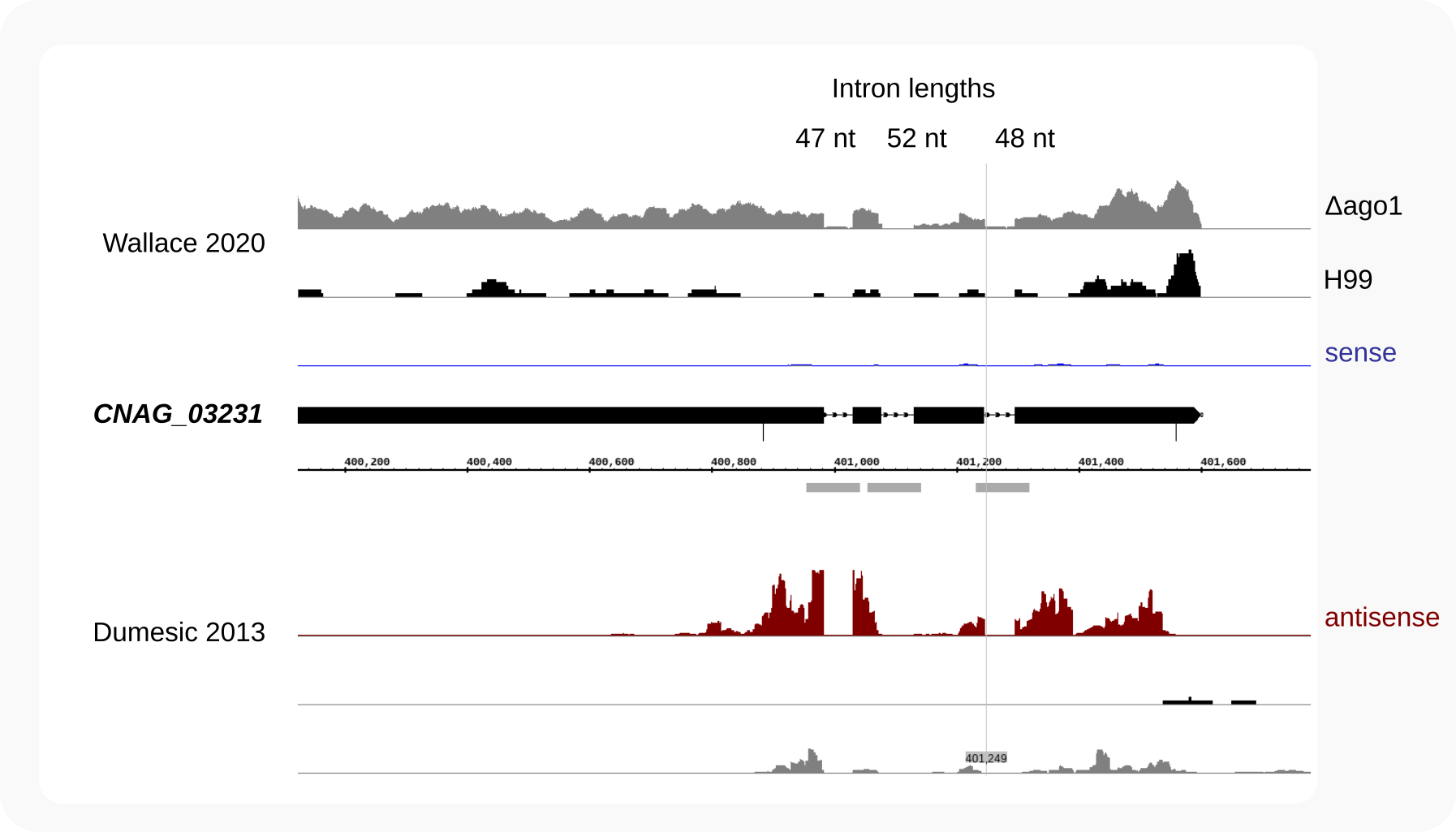
Figure 1. Introns in CNAG_03231 (CNAG_07721)¶
RNA-Seq [Wallace-2020] indicates the presence of three introns of 47, 52 and 48 nt in the primary transcript of CNAG_03231. Antisense siRNAs (dark red; [Dumesic-2013]) are mainly raised against regions not containing the intron sequences. The GTF reference (black bars) is from [Wallace-2020].
The CNAG_03231 region targeted by siRNAs contains three introns in the primary transcript. Zooming in to nucleotide resolution, provides detailed information on intron sequences and RNA alignments:
5’ flank |
length |
sequence |
3’ flank |
|---|---|---|---|
TGCTACTC |
47 |
GTGAGTACTTCTTTCGTGGCAGATCGTGAAACTCATTGCGCCATCAG |
CACTTTCT |
AGGTACAT |
52 |
GTACGTGTCTTTAGTCTTAGGATGAGCCTTAAAGCTGACGCCGCCTGCATAG |
TCGATCAA |
CACGCCCT |
48 |
GTGAGTCTGCACACGGTGATTCTAGTGGCCATGCTGATTGTATGGCAG |
CAACCCCT |
From the work directory Burke-2019/, access the cDNA reads for the wild type, as published by [Dumesic-2013]:
cd STAR-analysis0-h99_collapsed/SRR646636_collapsed_0mismatch-h99/
To find the number of unique cDNAs that cross above introns:
for i in 47 52 48; do samtools view samtoolsAligned.sortedByCoord.out.bam | egrep $'\t8\t40[01][0-9][0-9][0-9]' | grep -c ${i}N ; done[2]
The numbers for skipped introns of 47, 52 and 48 nt, respectively, parallel the relative heights of siRNA peaks observed at this locus:
22
5
28
To check the actual reads:
for i in 47 52 48; do samtools view samtoolsAligned.sortedByCoord.out.bam | egrep $'\t8\t40[01][0-9][0-9][0-9]' | grep ${i}N | less; done[2]
Most informative reads in the output are shown below; they adhere to siRNA characteristics typical for Cryptococcus, with a length of 20-24 nt [3] and a U as start-nucleotide.
Samtools analysis of 47N¶
Selected cDNAs [4] crossing the 47 nt gap in CNAG_03231:
218234_2 16 8 400963 255 22M47N2M * 0 0 AGTTTATCTGCACTTGCTACTCCA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
111057_6 16 8 400964 255 21M47N2M * 0 0 GTTTATCTGCACTTGCTACTCCA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
52775_2 16 8 400974 255 11M47N10M * 0 0 ACTTGCTACTCCACTTTCTGA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
304314_6 16 8 400974 255 11M47N11M * 0 0 ACTTGCTACTCCACTTTCTGAA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
34661_7 16 8 400977 255 8M47N15M * 0 0 TGCTACTCCACTTTCTGAATTCA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
45563_6 16 8 400977 255 8M47N16M * 0 0 TGCTACTCCACTTTCTGAATTCAA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
113319_20 16 8 400978 255 7M47N16M * 0 0 GCTACTCCACTTTCTGAATTCAA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
225966_9 16 8 400978 255 7M47N15M * 0 0 GCTACTCCACTTTCTGAATTCA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
95287_21 16 8 400979 255 6M47N16M * 0 0 CTACTCCACTTTCTGAATTCAA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
Samtools analysis of 52N¶
All found reads that skip the intron of 52 nt in CNAG_03231:
8613_8 16 8 401069 255 11M52N13M * 0 0 AGGAGGTACATTCGATCAATCTAA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
287820_4 16 8 401069 255 11M52N12M * 0 0 AGGAGGTACATTCGATCAATCTA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
791645_2 16 8 401070 255 10M52N13M * 0 0 GGAGGTACATTCGATCAATCTAA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
81818_1 16 8 401070 255 10M52N12M * 0 0 GGAGGTACATTCGATCAATCTA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
339877_2 16 8 401072 255 8M52N13M * 0 0 AGGTACATTCGATCAATCTAA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
Samtools analysis of 48N¶
Selection of reads that bridge the 48 nt intron in CNAG_03231:
103518_15 16 8 401232 255 16M48N9M * 0 0 CGGGACGCCACGCCCTCAACCCCTA * NH:i:1 HI:i:1 AS:i:25 nM:i:0
127466_8 16 8 401233 255 15M48N9M * 0 0 GGGACGCCACGCCCTCAACCCCTA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
194744_21 16 8 401234 255 14M48N9M * 0 0 GGACGCCACGCCCTCAACCCCTA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
9838_100 16 8 401235 255 13M48N9M * 0 0 GACGCCACGCCCTCAACCCCTA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
370126_1 16 8 401235 255 13M48N10M * 0 0 GACGCCACGCCCTCAACCCCTAC * NH:i:1 HI:i:1 AS:i:23 nM:i:0
183626_31 16 8 401236 255 12M48N9M * 0 0 ACGCCACGCCCTCAACCCCTA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
266556_14 16 8 401237 255 11M48N9M * 0 0 CGCCACGCCCTCAACCCCTA * NH:i:1 HI:i:1 AS:i:20 nM:i:0
154010_5 16 8 401238 255 10M48N9M * 0 0 GCCACGCCCTCAACCCCTA * NH:i:1 HI:i:1 AS:i:19 nM:i:0
Reads with 62N gaps enriched in rde1Δ¶
When Rde1 is absent, hardly any siRNA reads were associated with Ago1. Therefore, reads representing loci that still raise siRNAs in this strain will be most abundant. Among uncollapsed reads and also for cDNAs, those with a 62 nt intron stood out, which were mostly mapping to a unique locus. Analogous to the detailed analysis to identify loci producing reads with a 73 nt gap enriched in rde2Δ, we can select alignments with 62 nt gaps in cDNAs for rde1Δ_1. From the work directory Burke-2019/:
cd STAR-analysis0-h99_collapsed/SRR8697586_collapsed_0mismatch-h99/or, for rde1Δ_2cd STAR-analysis0-h99_collapsed/SRR8697587_collapsed_0mismatch-h99/
Of the cDNAs with ~60-70 nt gaps those with introns of 62 nt and 68 nt stand out [5]; the former are found by:
samtools view samtoolsAligned.sortedByCoord.out.bam | grep 62N | less
From the output, which is listed below for the first and second replicate,
it can be concluded that the reads map to chromosome 6, to a locus (previously) annotated as CNAG_07650. As shown in the IGB browser (Fig. 2, right panel and bottom), two other, intron-containg reads are to be expected, of 57 and 68 nt. Reads crossing these introns have also been detected, as described for selections with grep 57N and grep 68N below.
5’ flank |
length |
sequence |
3’ flank |
|---|---|---|---|
TCCTCTATG |
57 |
GTGAGTCGGACACCTCTTGCAAGTCATGGCTCACTCTCACTCGACATACGCTTTCAG |
TACCCAAGGGA |
GCTCTGTCT |
62 |
GTAAGCGGTCCACATTCACCGGATCTAAAAGGAGGTCGTACCAAGTACTGATGATAGGATAG |
ACATCCTTTTC |
CGATTTACA |
68 |
GTATGTTTAGCAGTACCCGTAATAATCTTATTGCGCTCACCACTTTCCATTCATCTTGCTGTGGGCAG |
GGAAAGACCTA |
In the absence of Ago1 (see the ago1Δ RNA-Seq trace), spliced transcripts become detectable for this pseudo-gene. This indicates that RNAi is directed against these transcripts, but, in view of the siRNA mapping, after splicing has been completed.
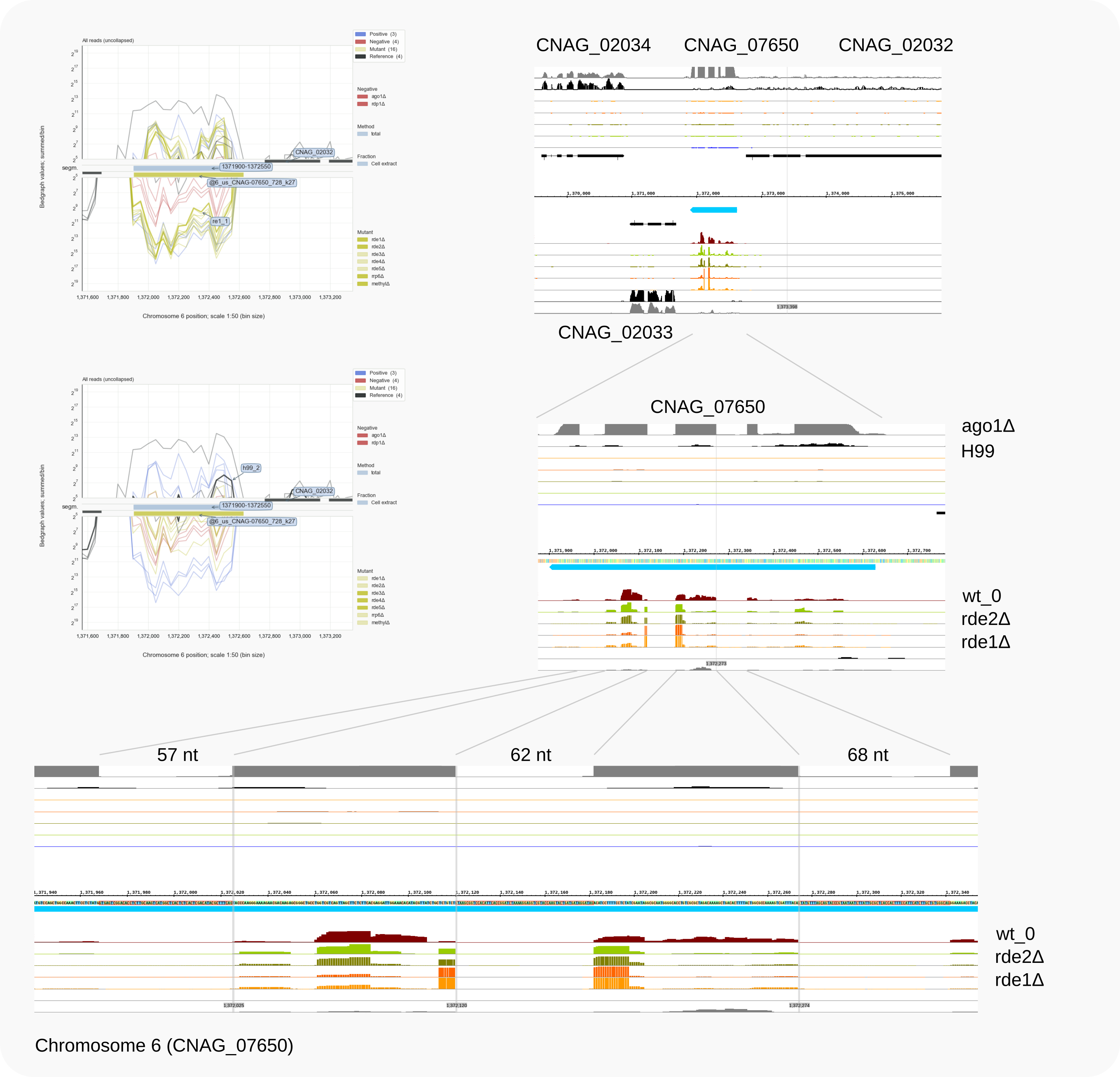
Figure 2. Reads across an 62 nt intron, relatively enriched in rde1Δ, mapped to CNAG_07650 on chr. 6.¶
ago1Δ) vs. wild type (H99).Return to the work directory:
cd ../..
Samtools analysis of rde1Δ_1¶
Check frequencies for reads bridging various intron-lengths.
62N.1¶
samtools view samtoolsAligned.sortedByCoord.out.bam | grep 62N | less
A selection of the output of this command, with reads bridging an intron of 62 nt, flanked by GCTCTGTCT at the 5’ splice site and by ACATCCTTTTC at the 3’ splice site:
4773_343 16 6 1372101 255 20M62N3M * 0 0 ATACGTTATCTGCTCTGTCTACA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
13090_91 0 6 1372102 255 19M62N4M * 0 0 TACGTTATCTGCTCTGTCTACAT * NH:i:1 HI:i:1 AS:i:23 nM:i:0
442_1876 16 6 1372102 255 19M62N3M * 0 0 TACGTTATCTGCTCTGTCTACA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
470_489 16 6 1372103 255 18M62N3M * 0 0 ACGTTATCTGCTCTGTCTACA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
7978_68 16 6 1372112 255 9M62N16M * 0 0 GCTCTGTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:25 nM:i:0
77907_141 16 6 1372113 255 8M62N16M * 0 0 CTCTGTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
151_16132 16 6 1372114 255 7M62N16M * 0 0 TCTGTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
81047_70 16 6 1372114 255 7M62N12M * 0 0 TCTGTCTACATCCTTTTCC * NH:i:1 HI:i:1 AS:i:19 nM:i:0
3738_1507 16 6 1372115 255 6M62N16M * 0 0 CTGTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
3112_2526 16 6 1372116 255 5M62N16M * 0 0 TGTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
2275_1351 16 6 1372118 3 3M62N21M * 0 0 TCTACATCCTTTTCCTCTATCGAA * NH:i:2 HI:i:2 AS:i:24 nM:i:0
9784_932 16 6 1372118 3 3M62N20M * 0 0 TCTACATCCTTTTCCTCTATCGA * NH:i:2 HI:i:1 AS:i:23 nM:i:0
2126_2300 16 6 1372119 3 2M62N21M * 0 0 CTACATCCTTTTCCTCTATCGAA * NH:i:2 HI:i:1 AS:i:23 nM:i:0
3967_416 16 6 1372119 3 2M62N20M * 0 0 CTACATCCTTTTCCTCTATCGA * NH:i:2 HI:i:2 AS:i:22 nM:i:0
68N.1¶
samtools view samtoolsAligned.sortedByCoord.out.bam | grep 68N | less
In transcripts from CNAG_07650 (no longer annotated as a coding gene) an intron of 68 nt flanked by, at the 5’ by CGATTTACA and at the 3’ end by GGAAAGACCTA, must have been spliced before siRNAs represented by the following reads were formed:
97642_19 16 6 1372254 255 21M68N4M * 0 0 GGCGCCAAAAGTCGATTTACAGGAA * NH:i:1 HI:i:1 AS:i:25 nM:i:0
12411_574 16 6 1372255 255 20M68N3M * 0 0 GCGCCAAAAGTCGATTTACAGGA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
26354_130 16 6 1372256 255 19M68N3M * 0 0 CGCCAAAAGTCGATTTACAGGA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
22610_261 16 6 1372257 255 18M68N4M * 0 0 GCCAAAAGTCGATTTACAGGAA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
42202_121 16 6 1372257 255 18M68N3M * 0 0 GCCAAAAGTCGATTTACAGGA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
26007_113 16 6 1372264 255 11M68N11M * 0 0 GTCGATTTACAGGAAAGACCTA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
7162_250 16 6 1372272 255 3M68N19M * 0 0 ACAGGAAAGACCTACACTTGGA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
18136_130 16 6 1372273 255 2M68N19M * 0 0 CAGGAAAGACCTACACTTGGA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
57N.1¶
Like the introns of 62 and 68 nt, the intron of 57 nt, flanked by TCCTCTATG at the 5’ end and by TACCCAAGGGA at the 3’ splice site, has been removed prior to the RNAi response:
samtools view samtoolsAligned.sortedByCoord.out.bam | grep 57N | less
30417_41 16 6 1371948 255 21M57N2M * 0 0 GCTGGCCAAACTTCCTCTATGTA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
36842_53 16 6 1371949 255 20M57N2M * 0 0 CTGGCCAAACTTCCTCTATGTA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
55878_55 16 6 1371950 255 19M57N2M * 0 0 TGGCCAAACTTCCTCTATGTA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
31807_109 16 6 1371951 255 18M57N2M * 0 0 GGCCAAACTTCCTCTATGTA * NH:i:1 HI:i:1 AS:i:20 nM:i:0
89005_50 16 6 1371951 255 18M57N6M * 0 0 GGCCAAACTTCCTCTATGTACCCA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
...
| 6536_201 0 13 730691 0 13M57N9M * 0 0 TGAATGATGAGGACTGGTACGG * NH:i:5 HI:i:1 AS:i:22 nM:i:0
| 11761_182 0 13 730691 0 13M57N10M * 0 0 TGAATGATGAGGACTGGTACGGA * NH:i:5 HI:i:1 AS:i:23 nM:i:0
| 19850_75 0 13 730691 0 13M57N8M * 0 0 TGAATGATGAGGACTGGTACG * NH:i:5 HI:i:1 AS:i:21 nM:i:0
| ...
| 30796_40 0 13 730695 0 9M57N12M * 0 0 TGATGAGGACTGGTACGGATG * NH:i:5 HI:i:1 AS:i:21 nM:i:0
| 41793_2 0 13 730695 0 9M57N15M * 0 0 TGATGAGGACTGGTACGGATGCTC * NH:i:5 HI:i:1 AS:i:24 nM:i:0
| 46264_208 0 13 730695 0 9M57N13M * 0 0 TGATGAGGACTGGTACGGATGC * NH:i:5 HI:i:1 AS:i:22 nM:i:0
| 130291_77 0 13 730695 0 9M57N14M * 0 0 TGATGAGGACTGGTACGGATGCT * NH:i:5 HI:i:1 AS:i:23 nM:i:0
Samtools analysis of rde1Δ_2¶
Frequencies for reads bridging various intron-lengths.
62N.2¶
samtools view samtoolsAligned.sortedByCoord.out.bam | grep 62N | less
65400_137 16 6 1372099 255 22M62N3M * 0 0 ACATACGTTATCTGCTCTGTCTACA * NH:i:1 HI:i:1 AS:i:25 nM:i:0
31284_67 16 6 1372100 255 21M62N3M * 0 0 CATACGTTATCTGCTCTGTCTACA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
6821_480 16 6 1372101 255 20M62N3M * 0 0 ATACGTTATCTGCTCTGTCTACA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
10029_70 0 6 1372102 255 19M62N4M * 0 0 TACGTTATCTGCTCTGTCTACAT * NH:i:1 HI:i:1 AS:i:23 nM:i:0
3076_2936 16 6 1372102 255 19M62N3M * 0 0 TACGTTATCTGCTCTGTCTACA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
4858_670 16 6 1372103 255 18M62N3M * 0 0 ACGTTATCTGCTCTGTCTACA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
19361_97 16 6 1372104 255 17M62N3M * 0 0 CGTTATCTGCTCTGTCTACA * NH:i:1 HI:i:1 AS:i:20 nM:i:0
18011_119 16 6 1372112 255 9M62N16M * 0 0 GCTCTGTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:25 nM:i:0
10588_248 16 6 1372113 255 8M62N16M * 0 0 CTCTGTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
962_21699 16 6 1372114 255 7M62N16M * 0 0 TCTGTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
33959_76 16 6 1372114 255 7M62N12M * 0 0 TCTGTCTACATCCTTTTCC * NH:i:1 HI:i:1 AS:i:19 nM:i:0
5502_2243 16 6 1372115 255 6M62N16M * 0 0 CTGTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
112_2737 16 6 1372116 255 5M62N16M * 0 0 TGTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
1057_72 16 6 1372117 255 4M62N16M * 0 0 GTCTACATCCTTTTCCTCTA * NH:i:1 HI:i:1 AS:i:20 nM:i:0
4642_449 16 6 1372117 255 4M62N20M * 0 0 GTCTACATCCTTTTCCTCTATCGA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
6487_319 16 6 1372117 255 4M62N21M * 0 0 GTCTACATCCTTTTCCTCTATCGAA * NH:i:1 HI:i:1 AS:i:25 nM:i:0
225_2659 16 6 1372118 3 3M62N21M * 0 0 TCTACATCCTTTTCCTCTATCGAA * NH:i:2 HI:i:1 AS:i:24 nM:i:0
5683_1719 16 6 1372118 3 3M62N20M * 0 0 TCTACATCCTTTTCCTCTATCGA * NH:i:2 HI:i:1 AS:i:23 nM:i:0
952_6646 16 6 1372119 3 2M62N21M * 0 0 CTACATCCTTTTCCTCTATCGAA * NH:i:2 HI:i:1 AS:i:23 nM:i:0
3604_13 16 6 1372119 3 2M62N6M1D14M * 0 0 CTACATCCTTTCCTCTATCGAA * NH:i:2 HI:i:2 AS:i:18 nM:i:0
4258_1039 16 6 1372119 3 2M62N20M * 0 0 CTACATCCTTTTCCTCTATCGA * NH:i:2 HI:i:2 AS:i:22 nM:i:0
44263_33 16 6 1372119 3 2M62N23M * 0 0 CTACATCCTTTTCCTCTATCGAATA * NH:i:2 HI:i:2 AS:i:25 nM:i:0
68N.2¶
samtools view samtoolsAligned.sortedByCoord.out.bam | grep 68N | less
51740_142 16 6 1372253 255 22M68N3M * 0 0 TGGCGCCAAAAGTCGATTTACAGGA * NH:i:1 HI:i:1 AS:i:25 nM:i:0
196_382 16 6 1372254 255 21M68N3M * 0 0 GGCGCCAAAAGTCGATTTACAGGA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
4076_581 16 6 1372255 255 20M68N3M * 0 0 GCGCCAAAAGTCGATTTACAGGA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
2919_168 16 6 1372256 255 19M68N3M * 0 0 CGCCAAAAGTCGATTTACAGGA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
1868_300 16 6 1372257 255 18M68N4M * 0 0 GCCAAAAGTCGATTTACAGGAA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
21005_103 16 6 1372257 255 18M68N3M * 0 0 GCCAAAAGTCGATTTACAGGA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
17473_84 16 6 1372271 255 4M68N19M * 0 0 TACAGGAAAGACCTACACTTGGA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
16493_371 16 6 1372272 255 3M68N19M * 0 0 ACAGGAAAGACCTACACTTGGA * NH:i:1 HI:i:1 AS:i:22 nM:i:0
57N.2¶
samtools view samtoolsAligned.sortedByCoord.out.bam | grep 57N | less
33192_109 16 6 1371951 255 18M57N2M * 0 0 GGCCAAACTTCCTCTATGTA * NH:i:1 HI:i:1 AS:i:20 nM:i:0
85469_51 16 6 1371951 255 18M57N6M * 0 0 GGCCAAACTTCCTCTATGTACCCA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
...
| 14662_190 0 13 730691 0 13M57N10M * 0 0 TGAATGATGAGGACTGGTACGGA * NH:i:5 HI:i:1 AS:i:23 nM:i:0
| 20227_215 0 13 730691 0 13M57N9M * 0 0 TGAATGATGAGGACTGGTACGG * NH:i:5 HI:i:1 AS:i:22 nM:i:0
| 22008_106 0 13 730691 0 13M57N8M * 0 0 TGAATGATGAGGACTGGTACG * NH:i:5 HI:i:1 AS:i:21 nM:i:0
| ...
| 4301_97 0 13 730695 0 9M57N14M * 0 0 TGATGAGGACTGGTACGGATGCT * NH:i:5 HI:i:1 AS:i:23 nM:i:0
| 13899_264 0 13 730695 0 9M57N13M * 0 0 TGATGAGGACTGGTACGGATGC * NH:i:5 HI:i:1 AS:i:22 nM:i:0
| 21925_46 0 13 730695 0 9M57N12M * 0 0 TGATGAGGACTGGTACGGATG * NH:i:5 HI:i:1 AS:i:21 nM:i:0
Reads with 73N gaps enriched in rde2Δ¶
For an analysis of alignments in the case of rde2Δ clone re2_1 (from the work directory Burke-2019/):
cd STAR-analysis0-h99_collapsed/SRR8697588_collapsed_0mismatch-h99/
Counting of intron-like gaps has been based on the presence of xxN within the cigar string for each alignment (in the 6th column in the alignments), where xx gives the number of skipped nucleotides to enable alignment of the two ends of the read. We can grep for this gap:
First check the number of candidates for multimappers (-c); 73 seems the right size for a peak [6]:
samtools view samtoolsAligned.sortedByCoord.out.bam | grep -c 73N
1689
Inspect the candidate reads that require a 73 nt gap to be aligned:
samtools view samtoolsAligned.sortedByCoord.out.bam | grep 73N | less
This will show many reads that align to single loci (especially around 108326-40 on chromosome 6). Because mainly multimappers were found to be enhanced that could bridge a 73 nt intron, we would like to filter the unique loci out. The single reads can be bypassed by an --invert-match, -v for unique mappers (NH:i:1\\s):
samtools view samtoolsAligned.sortedByCoord.out.bam | grep 73N | grep -v NH:i:1\\s | less
The loci in the output, from which a selection of abundant reads is reproduced, were also checked in the IGB genome browser. From these comparisons it can be concluded that:
Major loci where these reads map to are on chromosomes 1, 2, 3, 6, 7, 9, and 11.
These loci contain a element that is repeated in the reference genome:
once on chromosomes 1, 2, 3, 6 and 7;
twice on chr. 11 and
three times on chr. 9.
The repeated loci locate to the centromeres.
To some of these loci transcripts have been assigned that carry a 73 nt intron coinciding with the siRNA on the opposite strand.
locus |
NH [7] |
5’ flank |
sequence |
3’ flank |
|---|---|---|---|---|
6:108380 |
1 |
CCTGTTTTGCA |
GTAAGTAGCCAGTCAGCGGTGATTATTGTTGCGGCCTTTATCCTTCTTATAACGTTGACCCTTTAAGGCATAG |
AGTTCTCACT |
11:90550 |
7 |
AGGGTACTGAG |
GTGCGTTGTCGGCTGTATTCCCTTTTGACTTCTGGGTATGATGTCTGTGCTGACAGGTTGCGGCTTGATCTAG |
GTTCTTCGTC |
9:804640 |
1 |
GGGGTACTGAG |
GTGCGTTGTCAGCTGTATTCCCTTTTAACTTCTGGGTATGATGTCTGTGCTGACAGGTCGTGGCTTGATCTAG |
GTTCTTCGTC |
7:527210 |
1 |
AGGGTACTGAG |
GTGCGTTGTCGGCTGTATTCCCTTTTAACTTCTGGGTATGATGTCCATGCTGACAGGTTGCGGCTTGATCTAG |
GTTCTTCGTC |
6:818825 |
1 |
AGGGTACTGAG |
GTGCATTGTCGGCTGTATTCCCTTTTAACTTCTGGGTATGATGTCTGTGCTGACAGGTTGTGGCTTGATCTAG |
GTTCTTCGTC |
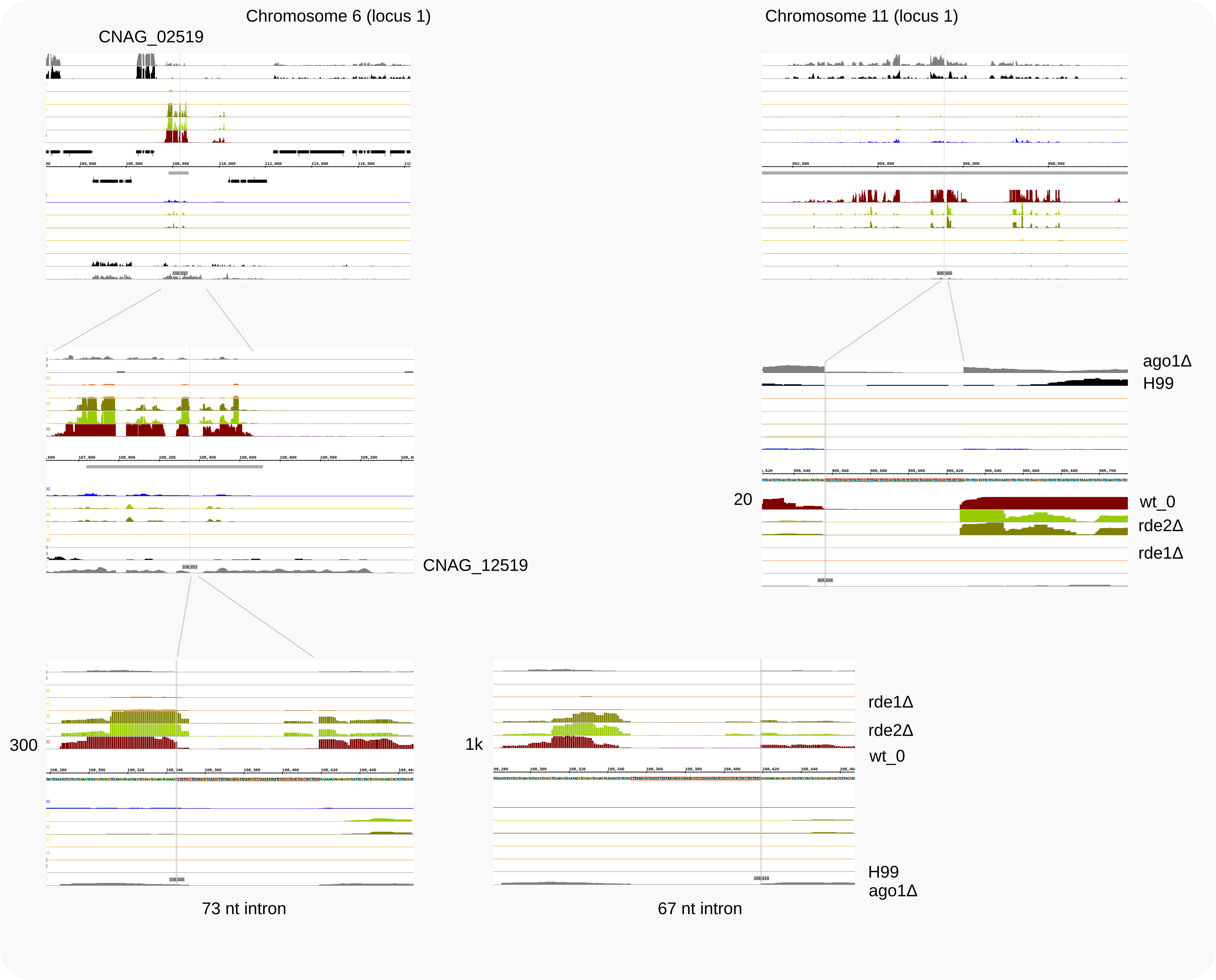
Figure 3. Reads mapped with 1 nt resolution bridging 73 nt introns¶
TCCGGGTCGAAGTGAGAACT--73N--TGCAAAACAGGA.AG bars should have been placed at the 5’ boundary: GGGTACTGAG--73N--GTTCTTCGTCTTCTTCA.While most reads with a 73 nucleotide gap map to centromeric repeat regions, one group of reads target ncRNA CNAG_12519, a unique locus (~108330-40) on the left arm of chromosome 6 nearby CNAG_02519 (Fig. 3 left panel). The population of siRNAs bridging a 73 nt gap appears to be minor in relation to the overall number of siRNAs generated for this locus. In view of the IGB alignments, this gap could refer to an intron spliced from a transcript on the same strand (opposite to CNAG_02519). Double-stranded RNA derived from sense-antisense transcription has been associated with siRNA synthesis in the presence of a core of maintained RNAi proteins [1] [Burroughs-2014]. In Cryptococcus, most siRNAs at a locus appear to be antisense to a transcript the level of which increases in the absence of RNAi (lane ago1Δ). Concomitantly, a less abundant group of siRNAs is found to be sense to this transcript. Thus, here we find that a less abundant siRNA could actually represent a spliced transcript from the same strand, while the majority of the same-stranded siRNAs are antisense to a transcript from the opposite strand. This observation suggests that RNAi related to double strandedness does not proceed in a symmetric manner. That is, there is preferential targetting of one transcript in the form of munro siRNAs, where two opposite transcripts are involved. This raises questions about the mechanism of siRNA generation.
RNA-transcripts from opposite strands, due to an unequal overlap of intronic regions that have been removed, will form imperfect double helices. Could the formation of double-strands interfere with transport or translation, with an RNAi response to overcome the problem as a result? Could the kind of bulges determine dicing? Or is the asymmetry linked to one transcript causing more problems, say during translation, than the one from the opposite strand?
The peak of multimapping cDNAs that could align by skipping the given gap of 73 nt in rde2Δ was the reason for looking at these introns. The reads associated with the repeated centromeric loci fit this observation.
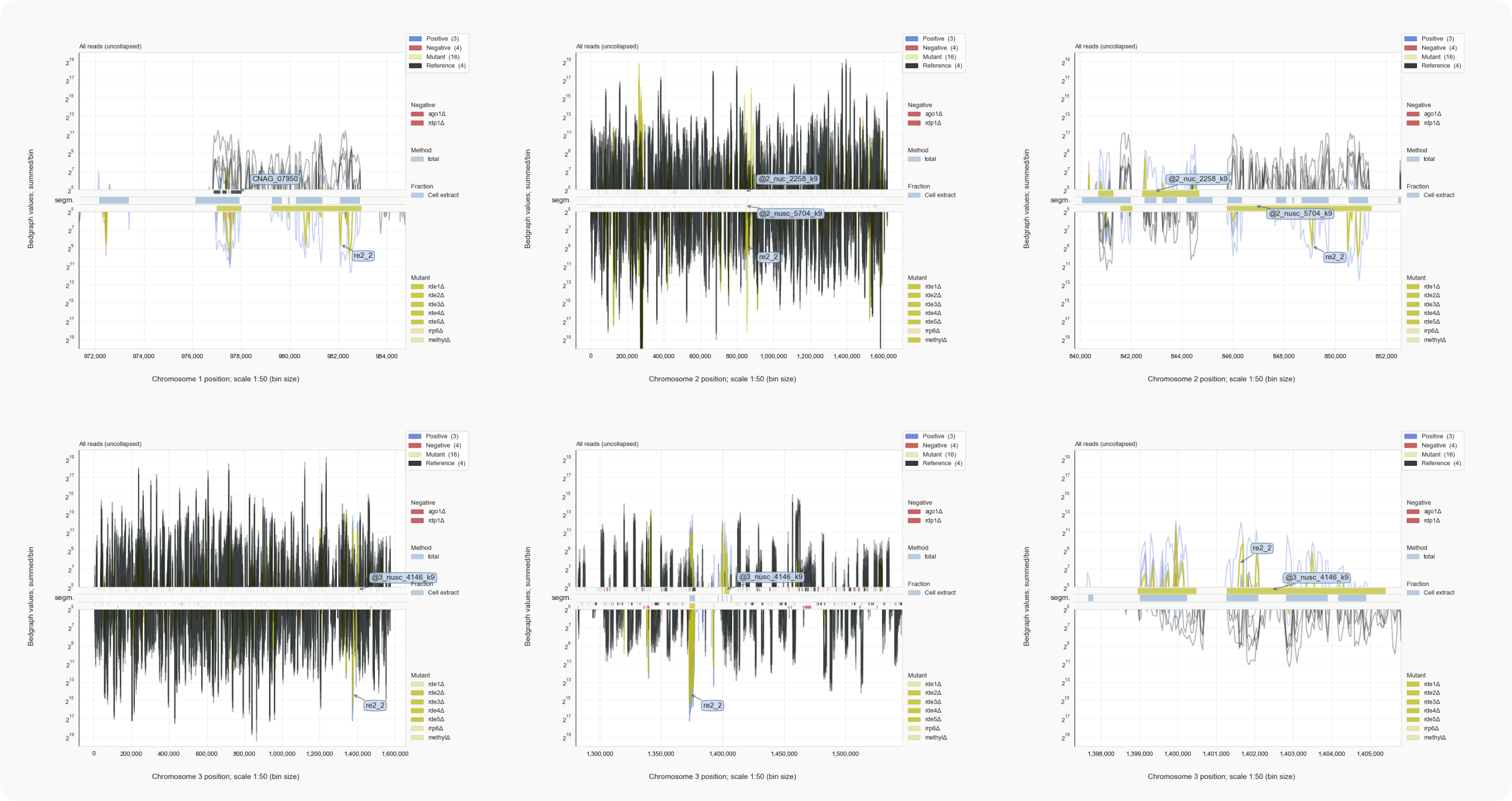
Figure 4A. C. neoformans loci with 73 nt introns in H99 rde2Δ.¶
Loci in h99_siRNAsegments.tsv with manually annotated features (olive-colored bar), were named like @3_nusc .. _k9, which indicated that the target transcript is on the opposite strand (@), is not uniq (nu), undergoes splicing (s) and is found within a centromeric region (c) that has been methylated on Lysine 9 of histone H3 (k9, [Dumesic-2015]).
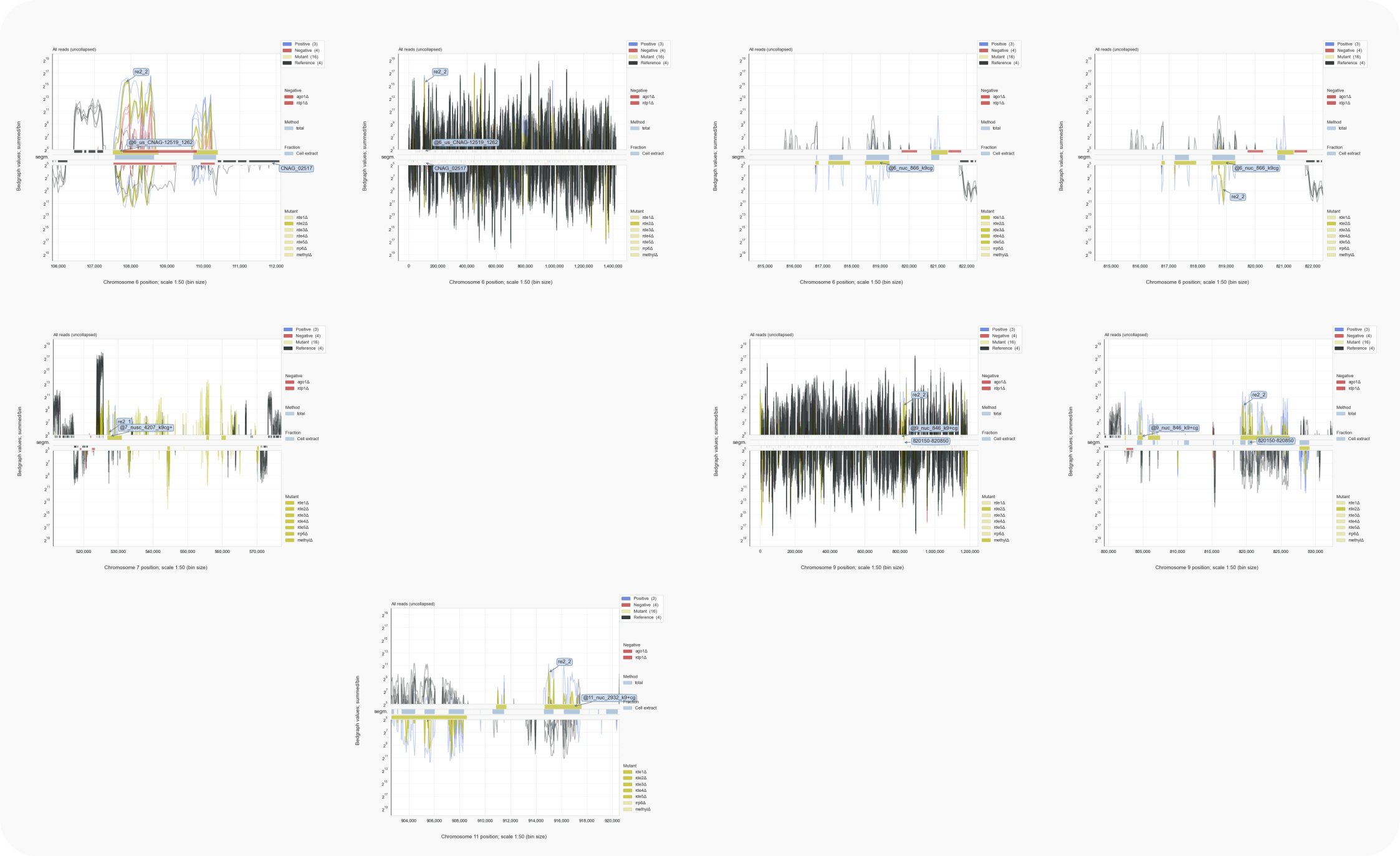
Figure 4B. C. neoformans loci with 73 nt introns in H99 rde2Δ (continued).¶
wt) and rde2Δ cells, but not for rde1,3,4,5Δ.The match between gene exons, i.e. spliced transcripts, and siRNAs targeting them, including those bridging a 73 nt intron, supports the idea that an RNAi response occurs downstream of splicing. If splicing in Cryptococcus is mostly co-transcriptional in line with findings for yeast and mammals, RNAi could be triggered during nuclear export of transcripts or when these are being translated by ribosomes.
Return to the work directory:
cd ../..
Samtools analysis of rde2Δ_1¶
Here, described in some detail, is a selection from the bam file that was in the grep-output.
| ...
| 958607_1 0 1 982384 0 21M73N7M * 0 0 GTTCGACTGGAGGGTACTGAGGTTCTTC * NH:i:9 HI:i:9 AS:i:28 nM:i:0
| 729686_2 16 1 982384 0 21M73N8M * 0 0 GTTCGACTGGAGGGTACTGAGGTTCTTCG * NH:i:9 HI:i:8 AS:i:29 nM:i:0
| 18098_113 0 1 982385 0 20M73N2M * 0 0 TTCGACTGGAGGGTACTGAGGT * NH:i:18 HI:i:7 AS:i:22 nM:i:0
| 38542_10 0 1 982385 0 20M73N3M * 0 0 TTCGACTGGAGGGTACTGAGGTT * NH:i:9 HI:i:6 AS:i:23 nM:i:0
| ...
| 1316670_1 16 1 982394 0 11M73N18M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCTTCAT * NH:i:9 HI:i:4 AS:i:29 nM:i:0
| 6948_51 16 1 982394 0 11M73N10M * 0 0 AGGGTACTGAGGTTCTTCGTC * NH:i:9 HI:i:6 AS:i:21 nM:i:0
| 34952_160 16 1 982394 0 11M73N8M * 0 0 AGGGTACTGAGGTTCTTCG * NH:i:9 HI:i:6 AS:i:19 nM:i:0
| 59419_184 16 1 982394 0 11M73N17M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:9 HI:i:7 AS:i:28 nM:i:0
| 77200_97 16 1 982394 0 11M73N11M * 0 0 AGGGTACTGAGGTTCTTCGTCT * NH:i:9 HI:i:5 AS:i:22 nM:i:0
| 255021_32 16 1 982394 0 11M73N12M * 0 0 AGGGTACTGAGGTTCTTCGTCTT * NH:i:9 HI:i:4 AS:i:23 nM:i:0
| ...
| 1449983_1 16 1 982395 0 10M73N10M * 0 0 GGGTACTGAGGTTCTTCGTC * NH:i:10 HI:i:5 AS:i:20 nM:i:0
| 71807_148 16 1 982395 0 10M73N17M * 0 0 GGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:5 AS:i:27 nM:i:0
| 215776_2 16 1 982395 0 10M73N21M * 0 0 GGGTACTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:7 AS:i:31 nM:i:0
| ...
| 840110_1 16 1 982399 0 6M73N16M * 0 0 ACTGAGGTTCTTCGTCTTCTTC * NH:i:10 HI:i:2 AS:i:22 nM:i:0
| 3061_1296 16 1 982400 0 5M73N17M * 0 0 CTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:8 AS:i:22 nM:i:0
| 22791_20 16 1 982400 0 5M73N21M * 0 0 CTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:9 AS:i:26 nM:i:0
| ...
| 1208513_2 16 1 982401 0 4M73N24M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCAATC * NH:i:10 HI:i:6 AS:i:28 nM:i:0
| 4888_238 16 1 982401 0 4M73N17M * 0 0 TGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:9 AS:i:21 nM:i:0
| 29430_206 16 1 982401 0 4M73N21M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:6 AS:i:25 nM:i:0
| 134302_32 16 1 982401 0 4M73N22M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:6 AS:i:26 nM:i:0
| 1112629_2 16 1 982401 0 4M73N19M * 0 0 TGAGGTTCTTCGTCTTCTTCATC * NH:i:10 HI:i:9 AS:i:23 nM:i:0
| 941_2130 16 1 982402 0 3M73N21M * 0 0 GAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:7 AS:i:24 nM:i:0
| 8093_136 16 1 982402 0 3M73N22M * 0 0 GAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:4 AS:i:25 nM:i:0
| 26122_49 16 1 982402 0 3M73N17M * 0 0 GAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:5 AS:i:20 nM:i:0
| 346340_1 16 1 982402 0 3M73N23M * 0 0 GAGGTTCTTCGTCTTCTTCATCCAAT * NH:i:10 HI:i:9 AS:i:26 nM:i:0
| 432628_3 16 1 982402 0 3M73N20M * 0 0 GAGGTTCTTCGTCTTCTTCATCC * NH:i:10 HI:i:4 AS:i:23 nM:i:0
| 483372_2 16 1 982402 0 3M73N24M * 0 0 GAGGTTCTTCGTCTTCTTCATCCAATC * NH:i:10 HI:i:2 AS:i:27 nM:i:0
| 1279985_2 16 1 982403 0 2M73N28M * 0 0 AGGTTCTTCGTCTTCTTCATCCAATCCTTG * NH:i:18 HI:i:8 AS:i:30 nM:i:0
| 145_17216 16 1 982403 0 2M73N22M * 0 0 AGGTTCTTCGTCTTCTTCATCCAA * NH:i:20 HI:i:10 AS:i:24 nM:i:0
| 547_52406 16 1 982403 0 2M73N21M * 0 0 AGGTTCTTCGTCTTCTTCATCCA * NH:i:20 HI:i:7 AS:i:23 nM:i:0
...
Note the entry 547_52406 at the end of the first column, which is the read name (see bam file description). After collapsing, the last digits provide the number of occurrences for individual cDNAs. High count numbers (like 52406, and further up 113, 160, 184, 97, 148, 1296, 238, 206, 2130, 136, 17216, 52406) for particular alignments point to a genomic region of interest, especially when other reads with a sequence overlap are found in the vicinity. Note the number of repeated mappings for these reads (12, 18, 9, 10, or 20); which is given by the 11th column, NH:i:xx. This is indicative for a repeated section and suitable for a transposon or remnants thereof. All these cDNAs were retrieved for the same expressed CNAG_07950 locus. The second column marks whether the read is on the plus (0) or minus (16) strand. The latter are antisense to CNAG_07950 transcripts, which is the expected strandedness for most siRNAs. In the collected alignments the ones that would not have been counted (because of a cigar string like: 8M1D11M73N2M or 10M1D11M73N2M) have been taken out.
The second region with significant hits is on chromosome 2:
| ...
| 1382464_1 0 2 850807 0 11M73N10M * 0 0 AGGGTACTGAGGTTCTTCGTC * NH:i:9 HI:i:1 AS:i:21 nM:i:0
| 44211_60 0 2 850807 0 11M73N11M * 0 0 AGGGTACTGAGGTTCTTCGTCT * NH:i:9 HI:i:7 AS:i:22 nM:i:0
| 58614_12 0 2 850807 0 11M73N12M * 0 0 AGGGTACTGAGGTTCTTCGTCTT * NH:i:9 HI:i:5 AS:i:23 nM:i:0
| 1316670_1 16 2 850807 0 11M73N18M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCTTCAT * NH:i:9 HI:i:8 AS:i:29 nM:i:0
| 6948_51 16 2 850807 0 11M73N10M * 0 0 AGGGTACTGAGGTTCTTCGTC * NH:i:9 HI:i:5 AS:i:21 nM:i:0
| 34952_160 16 2 850807 0 11M73N8M * 0 0 AGGGTACTGAGGTTCTTCG * NH:i:9 HI:i:2 AS:i:19 nM:i:0
| 59419_184 16 2 850807 0 11M73N17M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:9 HI:i:4 AS:i:28 nM:i:0
| 77200_97 16 2 850807 0 11M73N11M * 0 0 AGGGTACTGAGGTTCTTCGTCT * NH:i:9 HI:i:6 AS:i:22 nM:i:0
| 255021_32 16 2 850807 0 11M73N12M * 0 0 AGGGTACTGAGGTTCTTCGTCTT * NH:i:9 HI:i:2 AS:i:23 nM:i:0
| ...
| 1324043_1 16 2 850809 0 9M73N20M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCATCC * NH:i:10 HI:i:2 AS:i:29 nM:i:0
| 51981_235 16 2 850809 0 9M73N17M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:1 AS:i:26 nM:i:0
| 500596_5 16 2 850809 0 9M73N21M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:3 AS:i:30 nM:i:0
| ...
| 1386944_1 16 2 850812 0 6M73N14M * 0 0 ACTGAGGTTCTTCGTCTTCT * NH:i:10 HI:i:1 AS:i:20 nM:i:0
| 3025_622 16 2 850812 0 6M73N17M * 0 0 ACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:4 AS:i:23 nM:i:0
| 8699_1 16 2 850812 0 6M73N15M * 0 0 ACTGAGGTTCTTCGTCTTCTT * NH:i:10 HI:i:6 AS:i:21 nM:i:0
| 207702_26 16 2 850812 0 6M73N21M * 0 0 ACTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:2 AS:i:27 nM:i:0
| 815089_5 16 2 850812 0 6M73N22M * 0 0 ACTGAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:1 AS:i:28 nM:i:0
| 840110_1 16 2 850812 0 6M73N16M * 0 0 ACTGAGGTTCTTCGTCTTCTTC * NH:i:10 HI:i:4 AS:i:22 nM:i:0
| 3061_1296 16 2 850813 0 5M73N17M * 0 0 CTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:3 AS:i:22 nM:i:0
| 22791_20 16 2 850813 0 5M73N21M * 0 0 CTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:8 AS:i:26 nM:i:0
| ...
| 279985_2 16 2 850816 0 2M73N28M * 0 0 AGGTTCTTCGTCTTCTTCATCCAATCCTTG * NH:i:18 HI:i:9 AS:i:30 nM:i:0
| 145_17216 16 2 850816 0 2M73N22M * 0 0 AGGTTCTTCGTCTTCTTCATCCAA * NH:i:20 HI:i:9 AS:i:24 nM:i:0
| 547_52406 16 2 850816 0 2M73N21M * 0 0 AGGTTCTTCGTCTTCTTCATCCA * NH:i:20 HI:i:1 AS:i:23 nM:i:0
| 40968_35 16 2 850816 0 2M73N19M * 0 0 AGGTTCTTCGTCTTCTTCATC * NH:i:20 HI:i:3 AS:i:21 nM:i:0
| 90608_18 16 2 850816 0 2M73N24M * 0 0 AGGTTCTTCGTCTTCTTCATCCAATC * NH:i:20 HI:i:8 AS:i:26 nM:i:0
| ...
| 118691_29 16 2 850816 0 2M73N20M * 0 0 AGGTTCTTCGTCTTCTTCATCC * NH:i:20 HI:i:9 AS:i:22 nM:i:0
| 145062_54 16 2 850816 0 2M73N17M * 0 0 AGGTTCTTCGTCTTCTTCA * NH:i:20 HI:i:7 AS:i:19 nM:i:0
...
Other major loci where above reads map to are on chromosomes 3, 6, 7, 9, and 11:
| ...
| 448446_2 0 3 1401707 0 23M73N2M * 0 0 ATTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:8 AS:i:25 nM:i:0
| 145_17216 0 3 1401708 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:2 AS:i:24 nM:i:0
| 8093_136 0 3 1401708 0 22M73N3M * 0 0 TTGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:9 AS:i:25 nM:i:0
| 134302_32 0 3 1401708 0 22M73N4M * 0 0 TTGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:8 AS:i:26 nM:i:0
| 178341_2
| ...
| 547_52406 0 3 1401709 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:2 AS:i:23 nM:i:0
| 941_2130 0 3 1401709 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:3 AS:i:24 nM:i:0
| 22791_20 0 3 1401709 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:10 AS:i:26 nM:i:0
| 29430_206 0 3 1401709 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:4 AS:i:25 nM:i:0
| 45670_51 0 3 1401709 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:7 AS:i:28 nM:i:0
...
145006_2 0 6 108326 255 21M73N3M * 0 0 GTCCGGGTCGAAGTGAGAACTTGC * NH:i:1 HI:i:1 AS:i:24 nM:i:0
7730_1998 0 6 108327 255 20M73N3M * 0 0 TCCGGGTCGAAGTGAGAACTTGC * NH:i:1 HI:i:1 AS:i:23 nM:i:0
19324_191 0 6 108327 255 20M73N4M * 0 0 TCCGGGTCGAAGTGAGAACTTGCA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
24253_480 0 6 108327 3 20M73N2M * 0 0 TCCGGGTCGAAGTGAGAACTTG * NH:i:2 HI:i:2 AS:i:22 nM:i:0
198483_10 0 6 108327 255 20M73N9M * 0 0 TCCGGGTCGAAGTGAGAACTTGCAAAACA * NH:i:1 HI:i:1 AS:i:29 nM:i:0
...
905853_1 0 6 108331 255 16M73N12M * 0 0 GGTCGAAGTGAGAACTTGCAAAACAGGA * NH:i:1 HI:i:1 AS:i:28 nM:i:0
60442_179 0 6 108333 255 14M73N9M * 0 0 TCGAAGTGAGAACTTGCAAAACA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
149848_66 0 6 108333 255 14M73N8M * 0 0 TCGAAGTGAGAACTTGCAAAAC * NH:i:1 HI:i:1 AS:i:22 nM:i:0
167891_2 0 6 108333 255 14M73N6M * 0 0 TCGAAGTGAGAACTTGCAAA * NH:i:1 HI:i:1 AS:i:20 nM:i:0
...
1160351_1 16 6 108338 255 9M73N12M * 0 0 GTGAGAACTTGCAAAACAGGA * NH:i:1 HI:i:1 AS:i:21 nM:i:0
1466_794 0 6 108339 255 8M73N15M * 0 0 TGAGAACTTGCAAAACAGGAGGC * NH:i:1 HI:i:1 AS:i:23 nM:i:0
5973_7 0 6 108339 255 8M73N11M * 0 0 TGAGAACTTGCAAAACAGG * NH:i:1 HI:i:1 AS:i:19 nM:i:0
48071_263 0 6 108339 255 8M73N14M * 0 0 TGAGAACTTGCAAAACAGGAGG * NH:i:1 HI:i:1 AS:i:22 nM:i:0
92957_8 0 6 108339 255 8M73N18M * 0 0 TGAGAACTTGCAAAACAGGAGGCGTC * NH:i:1 HI:i:1 AS:i:26 nM:i:0
...
| 729686_2 16 6 818801 0 21M73N8M * 0 0 GTTCGACTGGAGGGTACTGAGGTTCTTCG * NH:i:9 HI:i:9 AS:i:29 nM:i:0
| 18098_113 0 6 818802 0 20M73N2M * 0 0 TTCGACTGGAGGGTACTGAGGT * NH:i:18 HI:i:9 AS:i:22 nM:i:0
| 38542_10 0 6 818802 0 20M73N3M * 0 0 TTCGACTGGAGGGTACTGAGGTT * NH:i:9 HI:i:9 AS:i:23 nM:i:0
| ...
| 1208513_2 16 6 818818 0 4M73N24M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCAATC * NH:i:10 HI:i:4 AS:i:28 nM:i:0
| 4888_238 16 6 818818 0 4M73N17M * 0 0 TGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:6 AS:i:21 nM:i:0
| 29430_206 16 6 818818 0 4M73N21M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:2 AS:i:25 nM:i:0
| 134302_32 16 6 818818 0 4M73N22M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:7 AS:i:26 nM:i:0
| 1112629_2 16 6 818818 0 4M73N19M * 0 0 TGAGGTTCTTCGTCTTCTTCATC * NH:i:10 HI:i:5 AS:i:23 nM:i:0
| 941_2130 16 6 818819 0 3M73N21M * 0 0 GAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:1 AS:i:24 nM:i:0
| 8093_136 16 6 818819 0 3M73N22M * 0 0 GAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:10 AS:i:25 nM:i:0
| 26122_49 16 6 818819 0 3M73N17M * 0 0 GAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:6 AS:i:20 nM:i:0
...
...
1030268_4 0 7 527202 0 22M73N7M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:8 AS:i:29 nM:i:0
547_52406 0 7 527203 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:9 AS:i:23 nM:i:0
941_2130 0 7 527203 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:9 AS:i:24 nM:i:0
22791_20 0 7 527203 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:2 AS:i:26 nM:i:0
29430_206 0 7 527203 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:3 AS:i:25 nM:i:0
45670_51 0 7 527203 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:4 AS:i:28 nM:i:0
207702_26 0 7 527203 0 21M73N6M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:1 AS:i:27 nM:i:0
...
620579_1 16 7 527206 0 18M73N4M * 0 0 ATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:6 AS:i:22 nM:i:0
3025_622 0 7 527207 0 17M73N6M * 0 0 TGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:2 AS:i:23 nM:i:0
3061_1296 0 7 527207 0 17M73N5M * 0 0 TGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:9 AS:i:22 nM:i:0
4888_238 0 7 527207 0 17M73N4M * 0 0 TGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:7 AS:i:21 nM:i:0
26122_49 0 7 527207 0 17M73N3M * 0 0 TGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:3 AS:i:20 nM:i:0
51981_235 0 7 527207 0 17M73N9M * 0 0 TGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:10 AS:i:26 nM:i:0
59419_184 0 7 527207 0 17M73N11M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:6 AS:i:28 nM:i:0
71807_148 0 7 527207 0 17M73N10M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCC * NH:i:10 HI:i:10 AS:i:27 nM:i:0
75411_29 0 7 527207 0 17M73N7M * 0 0 TGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:2 AS:i:24 nM:i:0
145062_54 0 7 527207 0 17M73N2M * 0 0 TGAAGAAGACGAAGAACCT * NH:i:20 HI:i:6 AS:i:19 nM:i:0
...
...
| 448446_2 0 9 804566 0 23M73N2M * 0 0 ATTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:10 AS:i:25 nM:i:0
| 145_17216 0 9 804567 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:3 AS:i:24 nM:i:0
| 8093_136 0 9 804567 0 22M73N3M * 0 0 TTGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:2 AS:i:25 nM:i:0
| 134302_32 0 9 804567 0 22M73N4M * 0 0 TTGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:1 AS:i:26 nM:i:0
| 815089_5 0 9 804567 0 22M73N6M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:2 AS:i:28 nM:i:0
| 827466_7 0 9 804567 0 22M73N5M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:1 AS:i:27 nM:i:0
| 1030268_4 0 9 804567 0 22M73N7M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:6 AS:i:29 nM:i:0
| 547_52406 0 9 804568 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:6 AS:i:23 nM:i:0
| 941_2130 0 9 804568 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:8 AS:i:24 nM:i:0
| 22791_20 0 9 804568 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:3 AS:i:26 nM:i:0
| 29430_206 0 9 804568 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:9 AS:i:25 nM:i:0
| 45670_51 0 9 804568 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:5 AS:i:28 nM:i:0
| 207702_26 0 9 804568 0 21M73N6M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:10 AS:i:27 nM:i:0
| ...
| 620579_1 16 9 804571 0 18M73N4M * 0 0 ATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:10 AS:i:22 nM:i:0
| 3025_622 0 9 804572 0 17M73N6M * 0 0 TGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:9 AS:i:23 nM:i:0
| 3061_1296 0 9 804572 0 17M73N5M * 0 0 TGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:1 AS:i:22 nM:i:0
| 4888_238 0 9 804572 0 17M73N4M * 0 0 TGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:1 AS:i:21 nM:i:0
| 26122_49 0 9 804572 0 17M73N3M * 0 0 TGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:2 AS:i:20 nM:i:0
| 51981_235 0 9 804572 0 17M73N9M * 0 0 TGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:3 AS:i:26 nM:i:0
| 71807_148 0 9 804572 0 17M73N10M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCC * NH:i:10 HI:i:9 AS:i:27 nM:i:0
| 75411_29 0 9 804572 0 17M73N7M * 0 0 TGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:6 AS:i:24 nM:i:0
| 145062_54 0 9 804572 0 17M73N2M * 0 0 TGAAGAAGACGAAGAACCT * NH:i:20 HI:i:2 AS:i:19 nM:i:0
...
448446_2 0 9 819503 0 23M73N2M * 0 0 ATTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:3 AS:i:25 nM:i:0
145_17216 0 9 819504 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:6 AS:i:24 nM:i:0
8093_136 0 9 819504 0 22M73N3M * 0 0 TTGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:1 AS:i:25 nM:i:0
134302_32 0 9 819504 0 22M73N4M * 0 0 TTGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:5 AS:i:26 nM:i:0
815089_5 0 9 819504 0 22M73N6M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:9 AS:i:28 nM:i:0
827466_7 0 9 819504 0 22M73N5M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:6 AS:i:27 nM:i:0
1030268_4 0 9 819504 0 22M73N7M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:2 AS:i:29 nM:i:0
547_52406 0 9 819505 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:8 AS:i:23 nM:i:0
941_2130 0 9 819505 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:10 AS:i:24 nM:i:0
22791_20 0 9 819505 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:1 AS:i:26 nM:i:0
29430_206 0 9 819505 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:7 AS:i:25 nM:i:0
45670_51 0 9 819505 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:9 AS:i:28 nM:i:0
207702_26 0 9 819505 0 21M73N6M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:6 AS:i:27 nM:i:0
...
620579_1 16 9 819508 0 18M73N4M * 0 0 ATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:9 AS:i:22 nM:i:0
3025_622 0 9 819509 0 17M73N6M * 0 0 TGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:1 AS:i:23 nM:i:0
3061_1296 0 9 819509 0 17M73N5M * 0 0 TGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:6 AS:i:22 nM:i:0
4888_238 0 9 819509 0 17M73N4M * 0 0 TGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:8 AS:i:21 nM:i:0
26122_49 0 9 819509 0 17M73N3M * 0 0 TGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:8 AS:i:20 nM:i:0
51981_235 0 9 819509 0 17M73N9M * 0 0 TGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:8 AS:i:26 nM:i:0
59419_184 0 9 819509 0 17M73N11M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:2 AS:i:28 nM:i:0
71807_148 0 9 819509 0 17M73N10M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCC * NH:i:10 HI:i:4 AS:i:27 nM:i:0
75411_29 0 9 819509 0 17M73N7M * 0 0 TGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:3 AS:i:24 nM:i:0
145062_54 0 9 819509 0 17M73N2M * 0 0 TGAAGAAGACGAAGAACCT * NH:i:20 HI:i:5 AS:i:19 nM:i:0
...
| 448446_2 0 9 820485 0 23M73N2M * 0 0 ATTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:6 AS:i:25 nM:i:0
| 145_17216 0 9 820486 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:5 AS:i:24 nM:i:0
| 8093_136 0 9 820486 0 22M73N3M * 0 0 TTGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:3 AS:i:25 nM:i:0
| 134302_32 0 9 820486 0 22M73N4M * 0 0 TTGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:2 AS:i:26 nM:i:0
| ...
| 1030268_4 0 9 820486 0 22M73N7M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:7 AS:i:29 nM:i:0
| 547_52406 0 9 820487 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:3 AS:i:23 nM:i:0
| 941_2130 0 9 820487 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:6 AS:i:24 nM:i:0
| 22791_20 0 9 820487 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:7 AS:i:26 nM:i:0
| 29430_206 0 9 820487 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:8 AS:i:25 nM:i:0
| 45670_51 0 9 820487 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:3 AS:i:28 nM:i:0
| 207702_26 0 9 820487 0 21M73N6M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:7 AS:i:27 nM:i:0
| ...
| 620579_1 16 9 820490 0 18M73N4M * 0 0 ATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:5 AS:i:22 nM:i:0
| 3025_622 0 9 820491 0 17M73N6M * 0 0 TGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:8 AS:i:23 nM:i:0
| 3061_1296 0 9 820491 0 17M73N5M * 0 0 TGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:7 AS:i:22 nM:i:0
| 4888_238 0 9 820491 0 17M73N4M * 0 0 TGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:5 AS:i:21 nM:i:0
| 26122_49 0 9 820491 0 17M73N3M * 0 0 TGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:9 AS:i:20 nM:i:0
| 51981_235 0 9 820491 0 17M73N9M * 0 0 TGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:9 AS:i:26 nM:i:0
| 59419_184 0 9 820491 0 17M73N11M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:1 AS:i:28 nM:i:0
| 71807_148 0 9 820491 0 17M73N10M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCC * NH:i:10 HI:i:6 AS:i:27 nM:i:0
| 75411_29 0 9 820491 0 17M73N7M * 0 0 TGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:7 AS:i:24 nM:i:0
| 145062_54 0 9 820491 0 17M73N2M * 0 0 TGAAGAAGACGAAGAACCT * NH:i:20 HI:i:9 AS:i:19 nM:i:0
| ...
| 58614_12 16 9 820496 0 12M73N11M * 0 0 AAGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:2 AS:i:23 nM:i:0
| 1324337_1 0 9 820497 0 11M73N16M * 0 0 AGACGAAGAACCTCAGTACCCTCCAGT * NH:i:9 HI:i:1 AS:i:27 nM:i:0
| 77200_97 0 9 820497 0 11M73N11M * 0 0 AGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:2 AS:i:22 nM:i:0
| 296263_1 0 9 820497 0 11M73N8M * 0 0 AGACGAAGAACCTCAGTAC * NH:i:10 HI:i:10 AS:i:19 nM:i:0
| ...
| 047965_1 16 9 820499 0 9M73N20M * 0 0 ACGAAGAACCTCAGTACCCTCCAGTCGAA * NH:i:9 HI:i:5 AS:i:29 nM:i:0
| 34952_160 0 9 820500 0 8M73N11M * 0 0 CGAAGAACCTCAGTACCCT * NH:i:9 HI:i:9 AS:i:19 nM:i:0
| 78116_1 0 9 820500 0 8M73N16M * 0 0 CGAAGAACCTCAGTACCCTCCAGT * NH:i:9 HI:i:8 AS:i:24 nM:i:0
| 113562_55 0 9 820500 0 8M73N14M * 0 0 CGAAGAACCTCAGTACCCTCCA * NH:i:9 HI:i:8 AS:i:22 nM:i:0
| 115290_16 0 9 820500 0 8M73N13M * 0 0 CGAAGAACCTCAGTACCCTCC * NH:i:9 HI:i:3 AS:i:21 nM:i:0
...
...
958607_1 0 11 905536 0 21M73N7M * 0 0 GTTCGACTGGAGGGTACTGAGGTTCTTC * NH:i:9 HI:i:8 AS:i:28 nM:i:0
729686_2 16 11 905536 0 21M73N8M * 0 0 GTTCGACTGGAGGGTACTGAGGTTCTTCG * NH:i:9 HI:i:4 AS:i:29 nM:i:0
18098_113 0 11 905537 0 20M73N2M * 0 0 TTCGACTGGAGGGTACTGAGGT * NH:i:18 HI:i:5 AS:i:22 nM:i:0
38542_10 0 11 905537 0 20M73N3M * 0 0 TTCGACTGGAGGGTACTGAGGTT * NH:i:9 HI:i:2 AS:i:23 nM:i:0
...
1316670_1 16 11 905546 0 11M73N18M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCTTCAT * NH:i:9 HI:i:9 AS:i:29 nM:i:0
6948_51 16 11 905546 0 11M73N10M * 0 0 AGGGTACTGAGGTTCTTCGTC * NH:i:9 HI:i:9 AS:i:21 nM:i:0
34952_160 16 11 905546 0 11M73N8M * 0 0 AGGGTACTGAGGTTCTTCG * NH:i:9 HI:i:5 AS:i:19 nM:i:0
59419_184 16 11 905546 0 11M73N17M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:9 HI:i:9 AS:i:28 nM:i:0
77200_97 16 11 905546 0 11M73N11M * 0 0 AGGGTACTGAGGTTCTTCGTCT * NH:i:9 HI:i:7 AS:i:22 nM:i:0
255021_32 16 11 905546 0 11M73N12M * 0 0 AGGGTACTGAGGTTCTTCGTCTT * NH:i:9 HI:i:6 AS:i:23 nM:i:0
294173_3 16 11 905546 0 11M73N14M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCT * NH:i:9 HI:i:5 AS:i:25 nM:i:0
...
1449983_1 16 11 905547 0 10M73N10M * 0 0 GGGTACTGAGGTTCTTCGTC * NH:i:10 HI:i:6 AS:i:20 nM:i:0
71807_148 16 11 905547 0 10M73N17M * 0 0 GGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:2 AS:i:27 nM:i:0
215776_2 16 11 905547 0 10M73N21M * 0 0 GGGTACTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:5 AS:i:31 nM:i:0
...
1324043_1 16 11 905548 0 9M73N20M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCATCC * NH:i:10 HI:i:8 AS:i:29 nM:i:0
51981_235 16 11 905548 0 9M73N17M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:5 AS:i:26 nM:i:0
500596_5 16 11 905548 0 9M73N21M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:5 AS:i:30 nM:i:0
...
840110_1 16 11 905551 0 6M73N16M * 0 0 ACTGAGGTTCTTCGTCTTCTTC * NH:i:10 HI:i:3 AS:i:22 nM:i:0
3061_1296 16 11 905552 0 5M73N17M * 0 0 CTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:10 AS:i:22 nM:i:0
22791_20 16 11 905552 0 5M73N21M * 0 0 CTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:6 AS:i:26 nM:i:0
...
1208513_2 16 11 905553 0 4M73N24M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCAATC * NH:i:10 HI:i:1 AS:i:28 nM:i:0
4888_238 16 11 905553 0 4M73N17M * 0 0 TGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:4 AS:i:21 nM:i:0
29430_206 16 11 905553 0 4M73N21M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:5 AS:i:25 nM:i:0
134302_32 16 11 905553 0 4M73N22M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:3 AS:i:26 nM:i:0
...
1279985_2 16 11 905555 0 2M73N28M * 0 0 AGGTTCTTCGTCTTCTTCATCCAATCCTTG * NH:i:18 HI:i:7 AS:i:30 nM:i:0
145_17216 16 11 905555 0 2M73N22M * 0 0 AGGTTCTTCGTCTTCTTCATCCAA * NH:i:20 HI:i:8 AS:i:24 nM:i:0
547_52406 16 11 905555 0 2M73N21M * 0 0 AGGTTCTTCGTCTTCTTCATCCA * NH:i:20 HI:i:10 AS:i:23 nM:i:0
40968_35 16 11 905555 0 2M73N19M * 0 0 AGGTTCTTCGTCTTCTTCATC * NH:i:20 HI:i:1 AS:i:21 nM:i:0
90608_18 16 11 905555 0 2M73N24M * 0 0 AGGTTCTTCGTCTTCTTCATCCAATC * NH:i:20 HI:i:6 AS:i:26 nM:i:0
118691_29 16 11 905555 0 2M73N20M * 0 0 AGGTTCTTCGTCTTCTTCATCC * NH:i:20 HI:i:2 AS:i:22 nM:i:0
145062_54 16 11 905555 0 2M73N17M * 0 0 AGGTTCTTCGTCTTCTTCA * NH:i:20 HI:i:4 AS:i:19 nM:i:0
...
| 448446_2 0 11 915039 0 23M73N2M * 0 0 ATTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:1 AS:i:25 nM:i:0
| 145_17216 0 11 915040 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:4 AS:i:24 nM:i:0
| 8093_136 0 11 915040 0 22M73N3M * 0 0 TTGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:8 AS:i:25 nM:i:0
| 134302_32 0 11 915040 0 22M73N4M * 0 0 TTGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:10 AS:i:26 nM:i:0
| ...
| 1030268_4 0 11 915040 0 22M73N7M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:10 AS:i:29 nM:i:0
| 547_52406 0 11 915041 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:5 AS:i:23 nM:i:0
| 941_2130 0 11 915041 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:4 AS:i:24 nM:i:0
| 22791_20 0 11 915041 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:5 AS:i:26 nM:i:0
| 29430_206 0 11 915041 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:1 AS:i:25 nM:i:0
| 45670_51 0 11 915041 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:1 AS:i:28 nM:i:0
| ...
| 620579_1 16 11 915044 0 18M73N4M * 0 0 ATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:2 AS:i:22 nM:i:0
| 3025_622 0 11 915045 0 17M73N6M * 0 0 TGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:10 AS:i:23 nM:i:0
| 3061_1296 0 11 915045 0 17M73N5M * 0 0 TGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:4 AS:i:22 nM:i:0
| 4888_238 0 11 915045 0 17M73N4M * 0 0 TGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:10 AS:i:21 nM:i:0
| 26122_49 0 11 915045 0 17M73N3M * 0 0 TGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:7 AS:i:20 nM:i:0
| 51981_235 0 11 915045 0 17M73N9M * 0 0 TGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:2 AS:i:26 nM:i:0
| 59419_184 0 11 915045 0 17M73N11M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:3 AS:i:28 nM:i:0
| 71807_148 0 11 915045 0 17M73N10M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCC * NH:i:10 HI:i:7 AS:i:27 nM:i:0
| 75411_29 0 11 915045 0 17M73N7M * 0 0 TGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:4 AS:i:24 nM:i:0
| 145062_54 0 11 915045 0 17M73N2M * 0 0 TGAAGAAGACGAAGAACCT * NH:i:20 HI:i:8 AS:i:19 nM:i:0
| ...
| 1047965_1 16 11 915053 0 9M73N20M * 0 0 ACGAAGAACCTCAGTACCCTCCAGTCGAA * NH:i:9 HI:i:4 AS:i:29 nM:i:0
| 34952_160 0 11 915054 0 8M73N11M * 0 0 CGAAGAACCTCAGTACCCT * NH:i:9 HI:i:4 AS:i:19 nM:i:0
| 78116_1 0 11 915054 0 8M73N16M * 0 0 CGAAGAACCTCAGTACCCTCCAGT * NH:i:9 HI:i:5 AS:i:24 nM:i:0
...
Samtools analysis of rde2Δ_2¶
Repeating this analysis for the second dataset for rde2Δ results in the same regions although with inequal read-abundance. For example, in the set from re2_1:
_1 3061_1296 16 1 982400 0 5M73N17M * 0 0 CTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:8 AS:i:22 nM:i:0
_1 547_52406 16 1 982403 0 2M73N21M * 0 0 AGGTTCTTCGTCTTCTTCATCCA * NH:i:20 HI:i:7 AS:i:23 nM:i:0
vs. the replicate re2_2:
_2 15789_1263 16 1 982400 0 5M73N17M * 0 0 CTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:2 AS:i:22 nM:i:0
_2 2888_21706 16 1 982403 0 2M73N21M * 0 0 AGGTTCTTCGTCTTCTTCATCCA * NH:i:20 HI:i:9 AS:i:23 nM:i:0
The relevant reads for re2_2 with skipped introns of 73 nt can be found by:
cd ../SRR8697589_collapsed_0mismatch-h99/samtools view samtoolsAligned.sortedByCoord.out.bam | grep 73N | less
| ...
| 34372_15 0 1 982385 0 20M73N7M * 0 0 TTCGACTGGAGGGTACTGAGGTTCTTC * NH:i:9 HI:i:2 AS:i:27 nM:i:0
| 136784_74 0 1 982385 0 20M73N2M * 0 0 TTCGACTGGAGGGTACTGAGGT * NH:i:18 HI:i:8 AS:i:22 nM:i:0
| ...
| 16049_111 16 1 982394 0 11M73N17M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:9 HI:i:6 AS:i:28 nM:i:0
| 80384_62 16 1 982394 0 11M73N11M * 0 0 AGGGTACTGAGGTTCTTCGTCT * NH:i:9 HI:i:1 AS:i:22 nM:i:0
| ...
| 6752_141 16 1 982396 0 9M73N17M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:3 AS:i:26 nM:i:0
| 12485_68 16 1 982396 0 9M73N21M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:6 AS:i:30 nM:i:0
| ...
| 6821_479 16 1 982399 0 6M73N17M * 0 0 ACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:2 AS:i:23 nM:i:0
| 31762_13 16 1 982399 0 6M73N22M * 0 0 ACTGAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:1 AS:i:28 nM:i:0
| 89661_21 16 1 982399 0 6M73N21M * 0 0 ACTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:9 AS:i:27 nM:i:0
| ...
| 15789_1263 16 1 982400 0 5M73N17M * 0 0 CTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:2 AS:i:22 nM:i:0
| ...
| 26160_142 16 1 982401 0 4M73N21M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:5 AS:i:25 nM:i:0
| 31989_145 16 1 982401 0 4M73N17M * 0 0 TGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:9 AS:i:21 nM:i:0
| 49569_19 16 1 982401 0 4M73N22M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:7 AS:i:26 nM:i:0
| 13657_1594 16 1 982402 0 3M73N21M * 0 0 GAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:8 AS:i:24 nM:i:0
| 32063_91 16 1 982402 0 3M73N22M * 0 0 GAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:7 AS:i:25 nM:i:0
| ...
| 1164_7585 16 1 982403 0 2M73N22M * 0 0 AGGTTCTTCGTCTTCTTCATCCAA * NH:i:20 HI:i:1 AS:i:24 nM:i:0
| 2888_21706 16 1 982403 0 2M73N21M * 0 0 AGGTTCTTCGTCTTCTTCATCCA * NH:i:20 HI:i:9 AS:i:23 nM:i:0
...
...
136784_74 0 2 850798 0 20M73N2M * 0 0 TTCGACTGGAGGGTACTGAGGT * NH:i:18 HI:i:2 AS:i:22 nM:i:0
...
16049_111 16 2 850807 0 11M73N17M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:9 HI:i:3 AS:i:28 nM:i:0
80384_62 16 2 850807 0 11M73N11M * 0 0 AGGGTACTGAGGTTCTTCGTCT * NH:i:9 HI:i:2 AS:i:22 nM:i:0
...
45603_121 16 2 850808 0 10M73N17M * 0 0 GGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:8 AS:i:27 nM:i:0
...
15789_1263 16 2 850813 0 5M73N17M * 0 0 CTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:4 AS:i:22 nM:i:0
...
26160_142 16 2 850814 0 4M73N21M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:6 AS:i:25 nM:i:0
31989_145 16 2 850814 0 4M73N17M * 0 0 TGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:1 AS:i:21 nM:i:0
49569_19 16 2 850814 0 4M73N22M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:6 AS:i:26 nM:i:0
13657_1594 16 2 850815 0 3M73N21M * 0 0 GAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:2 AS:i:24 nM:i:0
32063_91 16 2 850815 0 3M73N22M * 0 0 GAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:4 AS:i:25 nM:i:0
...
1164_7585 16 2 850816 0 2M73N22M * 0 0 AGGTTCTTCGTCTTCTTCATCCAA * NH:i:20 HI:i:7 AS:i:24 nM:i:0
2888_21706 16 2 850816 0 2M73N21M * 0 0 AGGTTCTTCGTCTTCTTCATCCA * NH:i:20 HI:i:7 AS:i:23 nM:i:0
...
...
| 1164_7585 0 3 1401708 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:8 AS:i:24 nM:i:0
| 31762_13 0 3 1401708 0 22M73N6M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:3 AS:i:28 nM:i:0
| 32063_91 0 3 1401708 0 22M73N3M * 0 0 TTGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:3 AS:i:25 nM:i:0
| ...
| 2888_21706 0 3 1401709 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:5 AS:i:23 nM:i:0
| 12076_30 0 3 1401709 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:3 AS:i:28 nM:i:0
| 12485_68 0 3 1401709 0 21M73N9M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:4 AS:i:30 nM:i:0
| 13657_1594 0 3 1401709 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:5 AS:i:24 nM:i:0
| 15333_11 0 3 1401709 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:6 AS:i:26 nM:i:0
| 26160_142 0 3 1401709 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:3 AS:i:25 nM:i:0
| ...
| 6752_141 0 3 1401713 0 17M73N9M * 0 0 TGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:6 AS:i:26 nM:i:0
| 6821_479 0 3 1401713 0 17M73N6M * 0 0 TGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:8 AS:i:23 nM:i:0
| 15789_1263 0 3 1401713 0 17M73N5M * 0 0 TGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:5 AS:i:22 nM:i:0
| 16049_111 0 3 1401713 0 17M73N11M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:2 AS:i:28 nM:i:0
| ...
| 31989_145 0 3 1401713 0 17M73N4M * 0 0 TGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:3 AS:i:21 nM:i:0
| 33757_4 0 3 1401713 0 17M73N15M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCTCCAG* NH:i:9 HI:i:1 AS:i:32 nM:i:0
| 45603_121 0 3 1401713 0 17M73N10M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCC * NH:i:10 HI:i:3 AS:i:27 nM:i:0
| ...
| 80384_62 0 3 1401719 0 11M73N11M * 0 0 AGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:9 AS:i:22 nM:i:0
...
...
1447_2229 0 6 108327 255 20M73N3M * 0 0 TCCGGGTCGAAGTGAGAACTTGC * NH:i:1 HI:i:1 AS:i:23 nM:i:0
14014_679 0 6 108327 3 20M73N2M * 0 0 TCCGGGTCGAAGTGAGAACTTG * NH:i:2 HI:i:1 AS:i:22 nM:i:0
21759_177 0 6 108327 255 20M73N4M * 0 0 TCCGGGTCGAAGTGAGAACTTGCA * NH:i:1 HI:i:1 AS:i:24 nM:i:0
...
68584_141 0 6 108333 255 14M73N9M * 0 0 TCGAAGTGAGAACTTGCAAAACA * NH:i:1 HI:i:1 AS:i:23 nM:i:0
...
11776_715 0 6 108339 255 8M73N15M * 0 0 TGAGAACTTGCAAAACAGGAGGC * NH:i:1 HI:i:1 AS:i:23 nM:i:0
25425_242 0 6 108339 255 8M73N14M * 0 0 TGAGAACTTGCAAAACAGGAGG * NH:i:1 HI:i:1 AS:i:22 nM:i:0
...
| 16049_111 16 6 818811 0 11M73N17M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:9 HI:i:9 AS:i:28 nM:i:0
| 80384_62 16 6 818811 0 11M73N11M * 0 0 AGGGTACTGAGGTTCTTCGTCT * NH:i:9 HI:i:3 AS:i:22 nM:i:0
| ...
| 45603_121 16 6 818812 0 10M73N17M * 0 0 GGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:2 AS:i:27 nM:i:0
| ...
| 6752_141 16 6 818813 0 9M73N17M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:5 AS:i:26 nM:i:0
| 12485_68 16 6 818813 0 9M73N21M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:5 AS:i:30 nM:i:0
| ...
| 6821_479 16 6 818816 0 6M73N17M * 0 0 ACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:5 AS:i:23 nM:i:0
| ...
| 15789_1263 16 6 818817 0 5M73N17M * 0 0 CTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:3 AS:i:22 nM:i:0
| ...
| 26160_142 16 6 818818 0 4M73N21M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:9 AS:i:25 nM:i:0
| 31989_145 16 6 818818 0 4M73N17M * 0 0 TGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:6 AS:i:21 nM:i:0
| 49569_19 16 6 818818 0 4M73N22M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:3 AS:i:26 nM:i:0
| 13657_1594 16 6 818819 0 3M73N21M * 0 0 GAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:9 AS:i:24 nM:i:0
| 32063_91 16 6 818819 0 3M73N22M * 0 0 GAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:5 AS:i:25 nM:i:0
| ...
| 1164_7585 16 6 818820 0 2M73N22M * 0 0 AGGTTCTTCGTCTTCTTCATCCAA * NH:i:20 HI:i:6 AS:i:24 nM:i:0
| 2888_21706 16 6 818820 0 2M73N21M * 0 0 AGGTTCTTCGTCTTCTTCATCCA * NH:i:20 HI:i:10 AS:i:23 nM:i:0
...
...
1164_7585 0 7 527202 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:5 AS:i:24 nM:i:0
...
2888_21706 0 7 527203 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:2 AS:i:23 nM:i:0
12076_30 0 7 527203 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:6 AS:i:28 nM:i:0
12485_68 0 7 527203 0 21M73N9M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:7 AS:i:30 nM:i:0
13657_1594 0 7 527203 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:4 AS:i:24 nM:i:0
15333_11 0 7 527203 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:1 AS:i:26 nM:i:0
26160_142 0 7 527203 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:2 AS:i:25 nM:i:0
...
6752_141 0 7 527207 0 17M73N9M * 0 0 TGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:8 AS:i:26 nM:i:0
6821_479 0 7 527207 0 17M73N6M * 0 0 TGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:10 AS:i:23 nM:i:0
15789_1263 0 7 527207 0 17M73N5M * 0 0 TGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:8 AS:i:22 nM:i:0
16049_111 0 7 527207 0 17M73N11M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:8 AS:i:28 nM:i:0
...
31989_145 0 7 527207 0 17M73N4M * 0 0 TGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:2 AS:i:21 nM:i:0
33757_4 0 7 527207 0 17M73N15M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCTCCAG* NH:i:9 HI:i:9 AS:i:32 nM:i:0
45603_121 0 7 527207 0 17M73N10M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCC * NH:i:10 HI:i:1 AS:i:27 nM:i:0
...
80384_62 0 7 527213 0 11M73N11M * 0 0 AGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:4 AS:i:22 nM:i:0
...
136784_74 16 7 527222 0 2M73N20M * 0 0 ACCTCAGTACCCTCCAGTCGAA * NH:i:18 HI:i:5 AS:i:22 nM:i:0
...
...
| 1164_7585 0 9 804567 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:3 AS:i:24 nM:i:0
| 31762_13 0 9 804567 0 22M73N6M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:10 AS:i:28 nM:i:0
| 32063_91 0 9 804567 0 22M73N3M * 0 0 TTGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:2 AS:i:25 nM:i:0
| ...
| 2888_21706 0 9 804568 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:8 AS:i:23 nM:i:0
| 12076_30 0 9 804568 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:2 AS:i:28 nM:i:0
| 12485_68 0 9 804568 0 21M73N9M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:1 AS:i:30 nM:i:0
| 13657_1594 0 9 804568 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:3 AS:i:24 nM:i:0
| 15333_11 0 9 804568 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:2 AS:i:26 nM:i:0
| 26160_142 0 9 804568 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:1 AS:i:25 nM:i:0
...
1164_7585 0 9 819504 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:9 AS:i:24 nM:i:0
31762_13 0 9 819504 0 22M73N6M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:7 AS:i:28 nM:i:0
32063_91 0 9 819504 0 22M73N3M * 0 0 TTGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:9 AS:i:25 nM:i:0
...
2888_21706 0 9 819505 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:3 AS:i:23 nM:i:0
12076_30 0 9 819505 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:7 AS:i:28 nM:i:0
12485_68 0 9 819505 0 21M73N9M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:10 AS:i:30 nM:i:0
13657_1594 0 9 819505 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:7 AS:i:24 nM:i:0
15333_11 0 9 819505 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:7 AS:i:26 nM:i:0
26160_142 0 9 819505 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:10 AS:i:25 nM:i:0
...
6752_141 0 9 819509 0 17M73N9M * 0 0 TGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:2 AS:i:26 nM:i:0
6821_479 0 9 819509 0 17M73N6M * 0 0 TGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:6 AS:i:23 nM:i:0
15789_1263 0 9 819509 0 17M73N5M * 0 0 TGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:7 AS:i:22 nM:i:0
16049_111 0 9 819509 0 17M73N11M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:7 AS:i:28 nM:i:0
18423_22 0 9 819509 0 17M73N8M * 0 0 TGAAGAAGACGAAGAACCTCAGTAC * NH:i:10 HI:i:7 AS:i:25 nM:i:0
27707_23 0 9 819509 0 17M73N7M * 0 0 TGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:1 AS:i:24 nM:i:0
31989_145 0 9 819509 0 17M73N4M * 0 0 TGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:4 AS:i:21 nM:i:0
...
45603_121 0 9 819509 0 17M73N10M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCC * NH:i:10 HI:i:4 AS:i:27 nM:i:0
...
80384_62 0 9 819515 0 11M73N11M * 0 0 AGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:8 AS:i:22 nM:i:0
...
| 1164_7585 0 9 820486 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:4 AS:i:24 nM:i:0
| 31762_13 0 9 820486 0 22M73N6M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:6 AS:i:28 nM:i:0
| 32063_91 0 9 820486 0 22M73N3M * 0 0 TTGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:6 AS:i:25 nM:i:0
| ...
| 2888_21706 0 9 820487 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:6 AS:i:23 nM:i:0
| 12076_30 0 9 820487 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:5 AS:i:28 nM:i:0
| 12485_68 0 9 820487 0 21M73N9M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:9 AS:i:30 nM:i:0
| 13657_1594 0 9 820487 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:1 AS:i:24 nM:i:0
| 15333_11 0 9 820487 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:3 AS:i:26 nM:i:0
| 26160_142 0 9 820487 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:8 AS:i:25 nM:i:0
| ...
| 6752_141 0 9 820491 0 17M73N9M * 0 0 TGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:1 AS:i:26 nM:i:0
| 6821_479 0 9 820491 0 17M73N6M * 0 0 TGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:1 AS:i:23 nM:i:0
| 15789_1263 0 9 820491 0 17M73N5M * 0 0 TGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:6 AS:i:22 nM:i:0
| 16049_111 0 9 820491 0 17M73N11M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:4 AS:i:28 nM:i:0
| ...
| 31989_145 0 9 820491 0 17M73N4M * 0 0 TGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:7 AS:i:21 nM:i:0
| 33757_4 0 9 820491 0 17M73N15M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCTCCAG* NH:i:9 HI:i:8 AS:i:32 nM:i:0
| 45603_121 0 9 820491 0 17M73N10M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCC * NH:i:10 HI:i:10 AS:i:27 nM:i:0
| ...
| 80384_62 0 9 820497 0 11M73N11M * 0 0 AGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:5 AS:i:22 nM:i:0
| ...
| 136784_74 16 9 820506 0 2M73N20M * 0 0 ACCTCAGTACCCTCCAGTCGAA * NH:i:18 HI:i:9 AS:i:22 nM:i:0
...
...
136784_74 0 11 905537 0 20M73N2M * 0 0 TTCGACTGGAGGGTACTGAGGT * NH:i:18 HI:i:7 AS:i:22 nM:i:0
...
16049_111 16 11 905546 0 11M73N17M * 0 0 AGGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:9 HI:i:5 AS:i:28 nM:i:0
80384_62 16 11 905546 0 11M73N11M * 0 0 AGGGTACTGAGGTTCTTCGTCT * NH:i:9 HI:i:6 AS:i:22 nM:i:0
...
45603_121 16 11 905547 0 10M73N17M * 0 0 GGGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:5 AS:i:27 nM:i:0
...
6752_141 16 11 905548 0 9M73N17M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:10 AS:i:26 nM:i:0
12485_68 16 11 905548 0 9M73N21M * 0 0 GGTACTGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:2 AS:i:30 nM:i:0
...
6821_479 16 11 905551 0 6M73N17M * 0 0 ACTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:7 AS:i:23 nM:i:0
...
15789_1263 16 11 905552 0 5M73N17M * 0 0 CTGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:9 AS:i:22 nM:i:0
...
26160_142 16 11 905553 0 4M73N21M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:4 AS:i:25 nM:i:0
31989_145 16 11 905553 0 4M73N17M * 0 0 TGAGGTTCTTCGTCTTCTTCA * NH:i:10 HI:i:5 AS:i:21 nM:i:0
49569_19 16 11 905553 0 4M73N22M * 0 0 TGAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:8 AS:i:26 nM:i:0
13657_1594 16 11 905554 0 3M73N21M * 0 0 GAGGTTCTTCGTCTTCTTCATCCA * NH:i:10 HI:i:10 AS:i:24 nM:i:0
32063_91 16 11 905554 0 3M73N22M * 0 0 GAGGTTCTTCGTCTTCTTCATCCAA * NH:i:10 HI:i:8 AS:i:25 nM:i:0
...
1164_7585 16 11 905555 0 2M73N22M * 0 0 AGGTTCTTCGTCTTCTTCATCCAA * NH:i:20 HI:i:10 AS:i:24 nM:i:0
2888_21706 16 11 905555 0 2M73N21M * 0 0 AGGTTCTTCGTCTTCTTCATCCA * NH:i:20 HI:i:4 AS:i:23 nM:i:0
...
| 1164_7585 0 11 915040 0 22M73N2M * 0 0 TTGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:2 AS:i:24 nM:i:0
| 31762_13 0 11 915040 0 22M73N6M * 0 0 TTGGATGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:5 AS:i:28 nM:i:0
| 32063_91 0 11 915040 0 22M73N3M * 0 0 TTGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:1 AS:i:25 nM:i:0
| ...
| 2888_21706 0 11 915041 0 21M73N2M * 0 0 TGGATGAAGAAGACGAAGAACCT * NH:i:20 HI:i:1 AS:i:23 nM:i:0
| 12076_30 0 11 915041 0 21M73N7M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTA * NH:i:10 HI:i:4 AS:i:28 nM:i:0
| 12485_68 0 11 915041 0 21M73N9M * 0 0 TGGATGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:3 AS:i:30 nM:i:0
| 13657_1594 0 11 915041 0 21M73N3M * 0 0 TGGATGAAGAAGACGAAGAACCTC * NH:i:10 HI:i:6 AS:i:24 nM:i:0
| 15333_11 0 11 915041 0 21M73N5M * 0 0 TGGATGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:5 AS:i:26 nM:i:0
| 26160_142 0 11 915041 0 21M73N4M * 0 0 TGGATGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:7 AS:i:25 nM:i:0
| ...
| 6752_141 0 11 915045 0 17M73N9M * 0 0 TGAAGAAGACGAAGAACCTCAGTACC * NH:i:10 HI:i:7 AS:i:26 nM:i:0
| 6821_479 0 11 915045 0 17M73N6M * 0 0 TGAAGAAGACGAAGAACCTCAGT * NH:i:10 HI:i:4 AS:i:23 nM:i:0
| 15789_1263 0 11 915045 0 17M73N5M * 0 0 TGAAGAAGACGAAGAACCTCAG * NH:i:10 HI:i:10 AS:i:22 nM:i:0
| 16049_111 0 11 915045 0 17M73N11M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:1 AS:i:28 nM:i:0
| ...
| 31989_145 0 11 915045 0 17M73N4M * 0 0 TGAAGAAGACGAAGAACCTCA * NH:i:10 HI:i:10 AS:i:21 nM:i:0
| 33757_4 0 11 915045 0 17M73N15M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCCTCCAG* NH:i:9 HI:i:7 AS:i:32 nM:i:0
| 45603_121 0 11 915045 0 17M73N10M * 0 0 TGAAGAAGACGAAGAACCTCAGTACCC * NH:i:10 HI:i:7 AS:i:27 nM:i:0
| ...
| 80384_62 0 11 915051 0 11M73N11M * 0 0 AGACGAAGAACCTCAGTACCCT * NH:i:9 HI:i:7 AS:i:22 nM:i:0
| ...
| 136784_74 16 11 915060 0 2M73N20M * 0 0 ACCTCAGTACCCTCCAGTCGAA * NH:i:18 HI:i:6 AS:i:22 nM:i:0
...