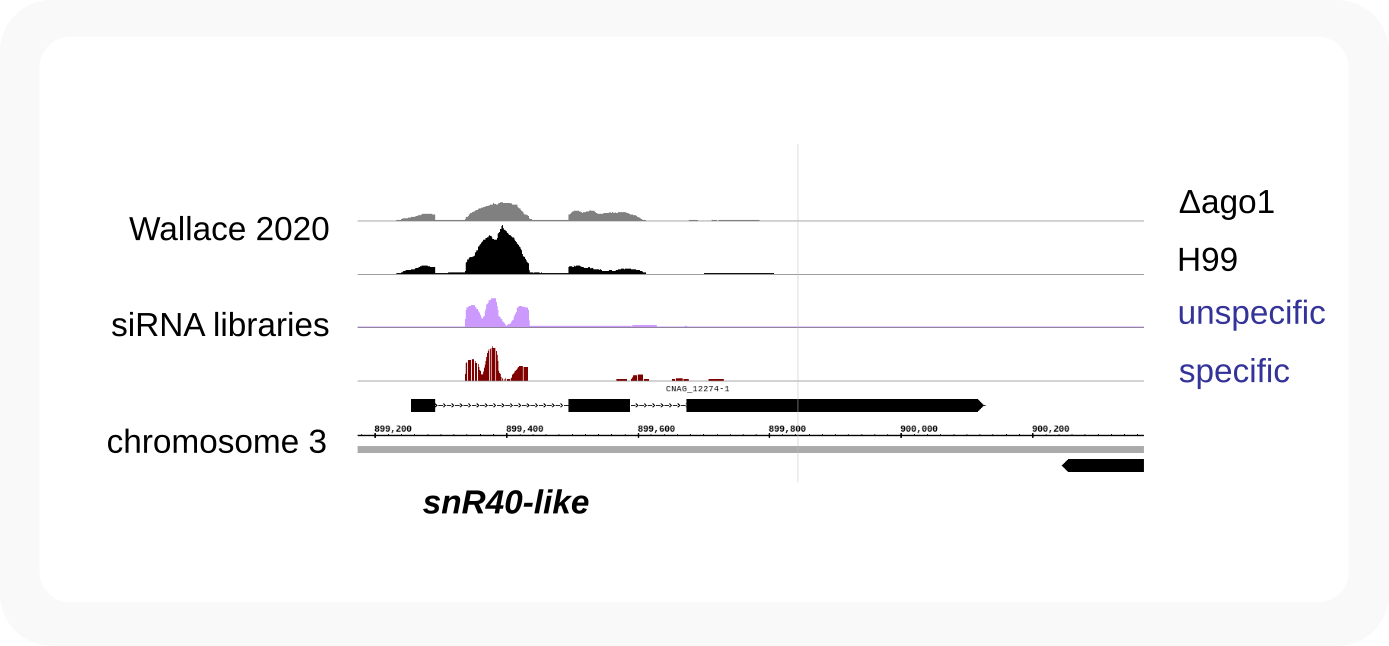
snR40-like¶
snR40-like_intronic-boxCDsnoRNA-97nt
Predicted target in 18S rRNA: (tgggtg)[g]tgGtgc[a]tggc (like snR40) or, uniq: (gggcaagt)ctgGtgccacag
Orthologue of yeast snR40
With canonical snR40 target in h34 of 18S rRNA the basepairing of the guide is not perfect. Full base-pairing is possible with another 18S region which has been documented as a second snR40 target in yeast 18S rRNA [Yang-2015].
Another snoRNA in Cryptococcus with snR40 specificity is available that also modifies the snR56 site on the opposite strand in h34.
Both guides of snR40 and snR40-like target the same nucleotide for modification but also overlap with one of the base-pairing sections of yeast snR4 that guides acetylation by Kre33 of C1280 in yeast, after modification by snR40 and prior to the event involving snR55 [Sharma-2017].
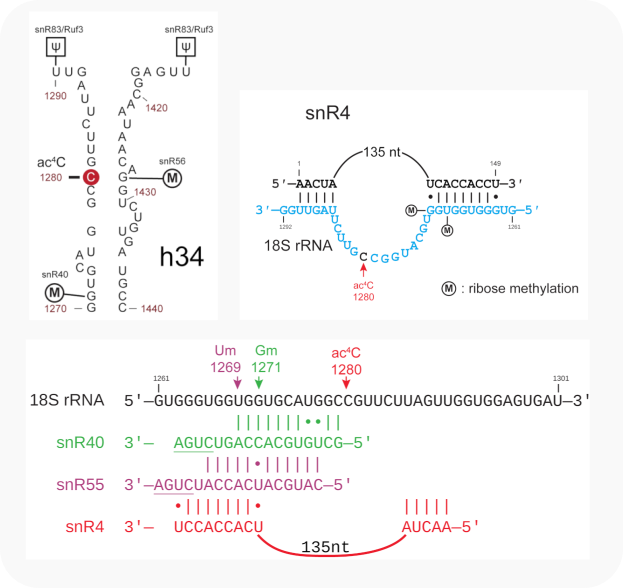
Overlapping base-pair interactions between h34 in 18S rRNA and snR40, snR4 and snR55 in yeast [from Sharma-2017]¶
Within an intron of a spliced non-coding transcript as CNAG_12274 (that possibly only consists of the one intron and first two exons as in JEC21)
C. neoformans snR40-like in Tremellomycetes
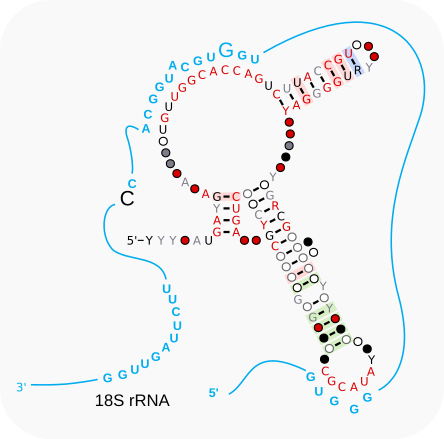
With only one guide linked to the D’ box and a large stemloop in between the C’ and D boxes, the secondary structure predicted for snR40-like differs from that of yeast snR40 [van.Nues-2011].
This stemloop contains a conserved motif (ATACGC) that might function as a protein binding site or as an extra guide although it does not appear to have a perfect match to the target of the accessory guide of yeast snR40 [van.Nues-2011].
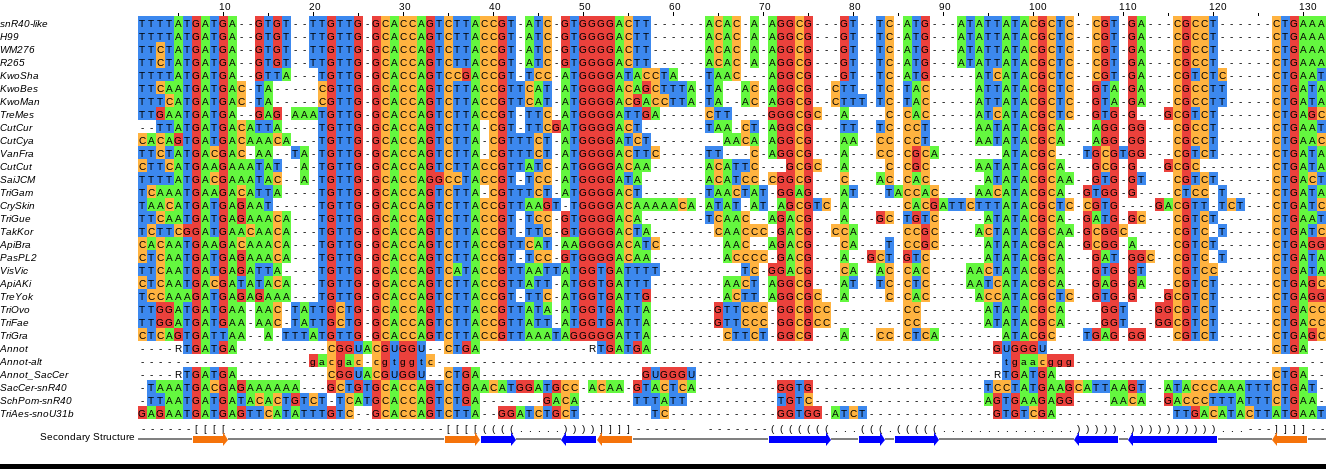
>AE017343.1:c1117380-1117285 Cryptococcus neoformans var. neoformans JEC21 chromosome 3 sequence TTTTATGATGAGTGTTTGTTGGCACCAGTCTTACCGTATCGTGGGGACTTACACAAGGCGGTTCATGATA TTATACGCTCCGTGACGCCTCTGAAA
Image source:
# STOCKHOLM 1.0
#=GF RW van Nues, June 2023
#=GF https://coalispr.codeberg.page/paper
#=GS snR40-like/1-97 DE intronic-boxCDsnoRNA-97nt ncrna 3:1117283:1117380:-1
snR40-like/1-97 TTTTATGATGAGTGT----TTGTTG-GCACCAGTCTTACCGT-ATC-GTGGGGACTT-----ACACA--AGGCG---GT--TC-ATG---ATATTATACGCTC--CGT-GA---CGCCT------CTGAAA
#=GS H99/111-899 DE CP003822.1:899230-900129 Cryptococcus neoformans var. grubii H99 chromosome 3, complete sequence
H99/111-899 TTTTATGATGAGTGT----TTGTTG-GCACCAGTCTTACCGT-ATC-GTGGGGACTT-----ACACA--AGGCG---GT--TC-ATG---ATATTATACGCTC--CGT-GA---CGCCT------CTGAAA
#=GS WM276/114-893 DE CP000288.1:c1085449-1084556 Cryptococcus gattii WM276 chromosome C, complete sequence
WM276/114-893 TTCTATGATGAGTGT----TTGTTG-GCACCAGTCTTACCGT-ATC-GTGGGGACTT-----ACACA--AGGCG---GT--TC-ATG---ATATTATACGCTC--CGT-GA---CGCCT------CTGAAA
#=GS R265/94-876 DE ENA|CP025759.1:1333206..1334501:ncRNA|CP025759.1:1333206..1334501:ncRNA.1 Cryptococcus gattii VGII R265 hypothetical RNA
R265/94-876 TTCTATGATGAGTGT----TTGTTG-GCACCAGTCTTACCGT-ATC-GTGGGGACTT-----ACACA--AGGCG---GT--TC-ATG---ATATTATACGCTC--CGT-GA---CGCCT------CTGAAA
#=GS KwoSha/5-102 DE NQVO01000013.1:461713-461814 Kwoniella shandongensis strain CBS 12478 scaffold00013, whole genome shotgun sequence
KwoSha/5-102 TTTTATGATGAGTTA-----TGTTG-GCACCAGTCCGACCGT-TCC-ATGGGGATACCT---ATAAC--AGGCG---GT--TC-ATG-----ATCATACGCTC--CGT-GA---CGTCTC-----CTGAAT
#=GS KwoBes/131-431 DE ASCK01000010.1:344013-344443 Kwoniella bestiolae CBS 10118 cont1.10, whole genome shotgun sequence
KwoBes/131-431 TTCAATGATGACTA------CGTTG-GCACCAGTCTTACCGTTCAT-ATGGGGACAGCTTT-ATAAC--AGGCG--CTT--TC-TAC-----ATTATACGCTC--GTA-GA---CGCCTT-----CTGATA
#=GS KwoMan/1-102 DE ASQE01000052.1:c395112-395011 Kwoniella mangroviensis CBS 8507 cont2.51, whole genome shotgun sequence
KwoMan/1-102 TTTCATGATGACTA------CGTTG-GCACCAGTCTTACCGTTCAT-ATGGGGACGACCTT-ATAAC--AGGCG--CTTT-TC-TAC-----ATTATACGCTC--GTA-GA---CGCCTT-----CTGATA
#=GS TreMes/97-394 DE AFVY01000049.1:c48481-48088 Tremella mesenterica DSM 1558 strain Fries TREMEscaffold_1_Cont49, whole genome shotgun sequence
TreMes/97-394 TTGAATGATGAGAGAAA---TGTTG-GCACCAGTCTTACCGT-TTC-ATGGGGATTG------ACTT--GGGCGC--A----C-CAC-----ATCATACGCTC--GTG-G---GCGTCT------CTGAGC
#=GS CutCur/1-92 DE NIUX01000033.1:c111158-111067 Cutaneotrichosporon curvatum strain ATCC 10567 Contig033, whole genome shotgun sequence
CutCur/1-92 --TTATGATGACATTA----TGTTG-GCACCAGTCTTA-CGT-TTCGATGGGGACTT------AACT--AGGCG---TT--TC-CCT-----AATATACGCA---AGG-GG---CGCCT------CTGAAT
#=GS CutCya/6-102 DE BEDZ01000041.1:c89956-89855 Cutaneotrichosporon cyanovorans DNA, scaffold: scaffold_41, strain: JCM_31833, whole genome shotgun sequence
CutCya/6-102 CACAGTGATGACAAACA---TGTTG-GCACCAGTCTTA-CGTTTCT-ATGGGGATCT------AACA--AGGCG---AA--CC-CCT-----AATATACGCA---AGG-GG---CGCCT------CTGAAC
#=GS VanFra/8-100 DE BEDY01000007.1:c340494-340395 Vanrija fragicola DNA, scaffold: scaffold_7, strain: JCM 1530, whole genome shotgun sequence
VanFra/8-100 TTCTATGACGACAATA----TGTTG-GCACCAGTCTTA-CGTTTCT-ATGGGGACTT------CTTC--AGGCG---A---CC-CGCA-------ATACGC---TGCGTGG---CGTCT------CTGATA
#=GS CutCut/4-97 DE JAMALK010000003.1:c630802-630706 Cutaneotrichosporon cutaneum strain P1411 ctg_3, whole genome shotgun sequence
CutCut/4-97 CTTCATGAAGAAATATA---TGTTG-GCACCAGTCTTACCGTTATC-ATGGGGACAA------ACAT--TCGCGC--A----C-CGC-----AATATACGCA---GCG-G---GCGC--------CTGATA
#=GS SaiJCM/10-105 DE BCLC01000002.1:c1094557-1094453 Saitozyma sp. JCM 24511 DNA, scaffold_1, whole genome shotgun sequence
SaiJCM/10-105 TTTTATGACGAAATACA---TGTTG-GCACCAGGCCTACCGT-TCC-ATGGGGATAA-----CATCC--CGGCG---C---AC-CAC------ATATACGCAA--GTG-GT---CGTCT------CTGACT
#=GS TriGam/7-101 DE BCJN01000009.1:c128968-128868 Trichosporon gamsii DNA, scaffold: scaffold_8, strain: JCM 9941, whole genome shotgun sequence
TriGam/7-101 TCAAATGAAGACATTA----TGTTG-GCACCAGTCTTA-CGTTTCT-ATGGGGACTT-----AACTAT--GGAG---AT---TACCAC----AACATACGCA--GTGG-G----CTCC-T-----CTGATA
#=GS CrySkin/8-118 DE BCHT01000003.1:88030-88147 Cryptococcus skinneri DNA, scaffold: scaffold_2, strain: JCM 9039, whole genome shotgun sequence
CrySkin/8-118 TAACATGATGAGAA-----TTGTTG-GCACCAGTCTTACCGTTAAGT-TGGGGACAAAAACAATATAT--AGCGTC-A------CACGATTCTTTATACGCTC-CGTG----GACGTT-TCT---CTGATC
#=GS TriGue/6-104 DE BCJX01000002.1:c2417929-2417826 Trichosporon guehoae DNA, scaffold: scaffold_1, strain: JCM 10690, whole genome shotgun sequence
TriGue/6-104 TTCAATGATGAGAAACA---TGTTG-GCACCAGTCTTACCGT-TCC-GTGGGGACAT------CAAC--AGACG---A---GC-TGTC-----ATATACGCA--GATG-GC---CGTCT------CTGAAT
#=GS TakKor/4-103 DE BCKT01000003.1:c1832418-1832316 Takashimella koratensis DNA, scaffold: scaffold_2, strain: JCM 12878, whole genome shotgun sequence
TakKor/4-103 TCTTCGGATGAACAACA---TGTTG-GCACCAGTCTTACCGT-TTC-GTGGGGACTA-----CAACCC--GACG--CCA-----CCGC----ACTATACGCAA-GCGGC-----CGTC-T-----CTGATC
#=GS ApiBra/7-103 DE JAMALJ010000047.1:575-677 Apiotrichum brassicae strain M2204 ctg_47, whole genome shotgun sequence
ApiBra/7-103 CACAATGAAGACAAACA---TGTTG-GCACCAGTCTTACCGTTCAT-AAGGGGACATC------AAC--AGACG---CA---T-CCGC-----ATATACGCA--GCGG-A----CGTCT------CTGAGG
#=GS PasPL2/7-103 DE JAMFRE010000001.1:c714444-714342 Pascua sp. PL2904B ctg_1, whole genome shotgun sequence
PasPL2/7-103 CTCAATGATGAGAAACA---TGTTG-GCACCAGTCTTACCGT-TCC-GTGGGGACAA------ACCCC--GACG---A--GCT-GTC------ATATACGCA---GAT-GGC--CGTC-T-----CTGATA
#=GS VisVic/7-106 DE JADPYG010000088.1:4268340-4268445 Vishniacozyma victoriae isolate T18_1_22C Contig_88, whole genome shotgun sequence
VisVic/7-106 TTCAATGATGAGATTA----TGTTG-GCACCAGTCATACCGTTAATTATGGTGATTTT-------TC--GGACG---CA--AC-CAC----AACTATACGCA---GTG-GT---CGTCC------CTGATA
#=GS ApiAKi/7-105 DE PQXP01000106.1:c41595-41491 Apiotrichum akiyoshidainum strain HP2023 Contig878, whole genome shotgun sequence
ApiAKi/7-105 CTCAATGACGATATACA---TGTTG-GCACCAGTCTTACCGTTATT-ATGGTGATTT------AACT--AGGCG---AT--TC-CTC----AATCATACGCA---GAG-GA---CGTCT------CTGAGC
#=GS TreYok/9-103 DE BRDC01000038.1:c226795-226693 Tremella yokohamensis NBRC 100148 DNA, KCNB35TY.38, whole genome shotgun sequence
TreYok/9-103 TCCAAAGATGAGAGAAA---TGTTG-GCACCAGTCTTACCGT-TTC-ATGGTGATTG------ACTT--AGGCGC--A----C-CAC-----ACCATACGCTC--GTG-G---GCGTCT------CTGAGG
#=GS TriOvo/6-99 DE JXYN01000008.1:999480-999578 Trichosporon ovoides strain JCM 9940 scaffold_0008, whole genome shotgun sequence
TriOvo/6-99 TTGGATGATGAAAACTA--TTGCTG-GCACCAGTCTTACCGTTATA-ATGGTGATTA-----GTTCCC--GGCGCC--------CC-------ATATACGCA----GGT---GGCGTCT------CTGACC
#=GS TriFae/2-97 DE JXYK01000001.1:c4469522-4469426 Trichosporon faecale strain JCM 2941 scaffold_0001, whole genome shotgun sequence
TriFae/2-97 TTGGATGATGAAAACTA--TTGCTG-GCACCAGTCTTACCGTTATT-ATGGTGATTA-----GTTCCC--GGCGCC--------CC-------ATATACGCA----GGT---GGCGTCT------CTGACC
#=GS TriGra/11-105 DE BCJO01000005.1:466968-467072 Trichosporon gracile DNA, scaffold: scaffold_4, strain: JCM 10018, whole genome shotgun sequence
TriGra/11-105 CTCAGTGATTAAATTTA---TGTTG-GCACCAGTCTTACCGTTAAATAGGGGGATTA------CTTCT--GGCG---A---CC-CTCA-------ATACGC---TGAG-GG---CGTCT------CTGAGC
Annot/7-47 ----RTGATGA----------CGGUACGUGGU--CTGA------------RTGATGA-------------------------------------GUGGGU-------------------------CTGA--
Annot-alt/2-22 -------------------gacgac-cgtggtc--------------------------------------------------------------tgaacggg----------------------------
Annot_SacCer/7-45 ----RTGATGA----------CGGUACGUGGU--CTGA------------------GUGGGU--------------------------------RTGATGA------------------------CTGA--
SacCer-snR40/1-95 -TAAATGACGAGAAAAAA---GCTGTGCACCAGTCTGAACATGGATGCC-ACAA-GTACTCA--------GGTG-------------T------CCTATGAAGCATTAAGT--ATACCCAAATTTCTGAT-
SchPom-snR40/1-81 -TTAATGATGATACACTGTCT-TCATGCACCAGTCTGA-------GACA------TTTATT---------TGTC-------------A------GTGAAGAGG----AACA--GACCCTTTATTTCTGAA-
#=GC SS_cons --------------------------------------((((.....))))------------------(((((((...(((.(((((...............))))).))))))))))...---------
//