SRP RNA¶
The signal recognition particle (SRP) is an evolutionarily conserved RNP involved in the export of proteins to the endoplasmic reticulum (ER). During this targeting the signalpeptide at the N-terminus of the nascent protein is recognized by Srp54 at the peptide exit tunnel of the ribosome. Meanwhile translation is arrested by interaction of Srp14 with the peptidyl transferase center. The distance between these sites on the ribosome is bridged by the extended and kinked shape of the RNA component of the particle. The SRP RNA (also referred to as 7SL, 6S, ffs, scR1, or 4.5S RNA) folds into an extended helix with two domains at either end. The middle section of the primary structure forms one of these domains, the S-domain, with helices 6 and 8 containing conserved structural elements that are bound by Srp19 (Sec65) and Srp54. The apical ends of these helices are brought together by Srp19 interacting with an adenosine in the loop of helix 6 and base interactions between both loops. The 5’ and 3’ ends of the RNA assemble into the Alu-domain where Srp14 and Srp9 (Srp21) associate, presumably by means of a highly conserved GUAAU sequence near the 5’ end [Strub-1999]. The rod-like stem connecting the Alu and S domains contains a conserved region (5e) marked by a bulge that has been found to genetically interact with Srp72 [van.Nues-2008], which forms a complex with Srp68.
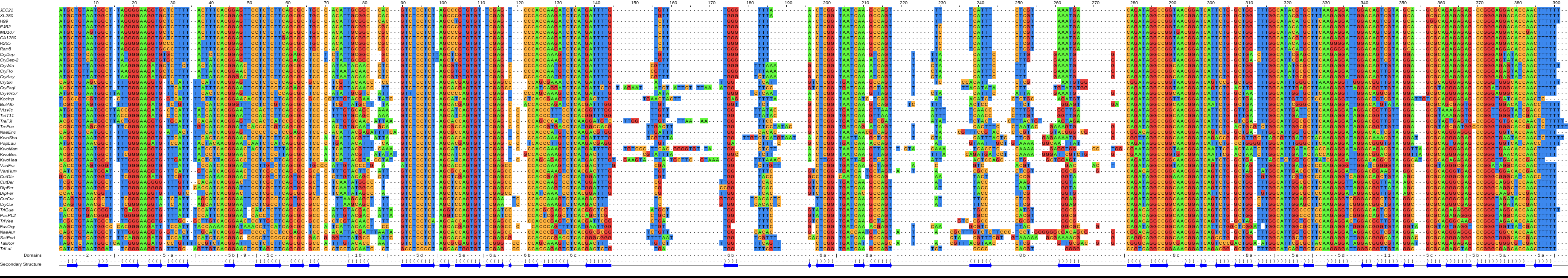
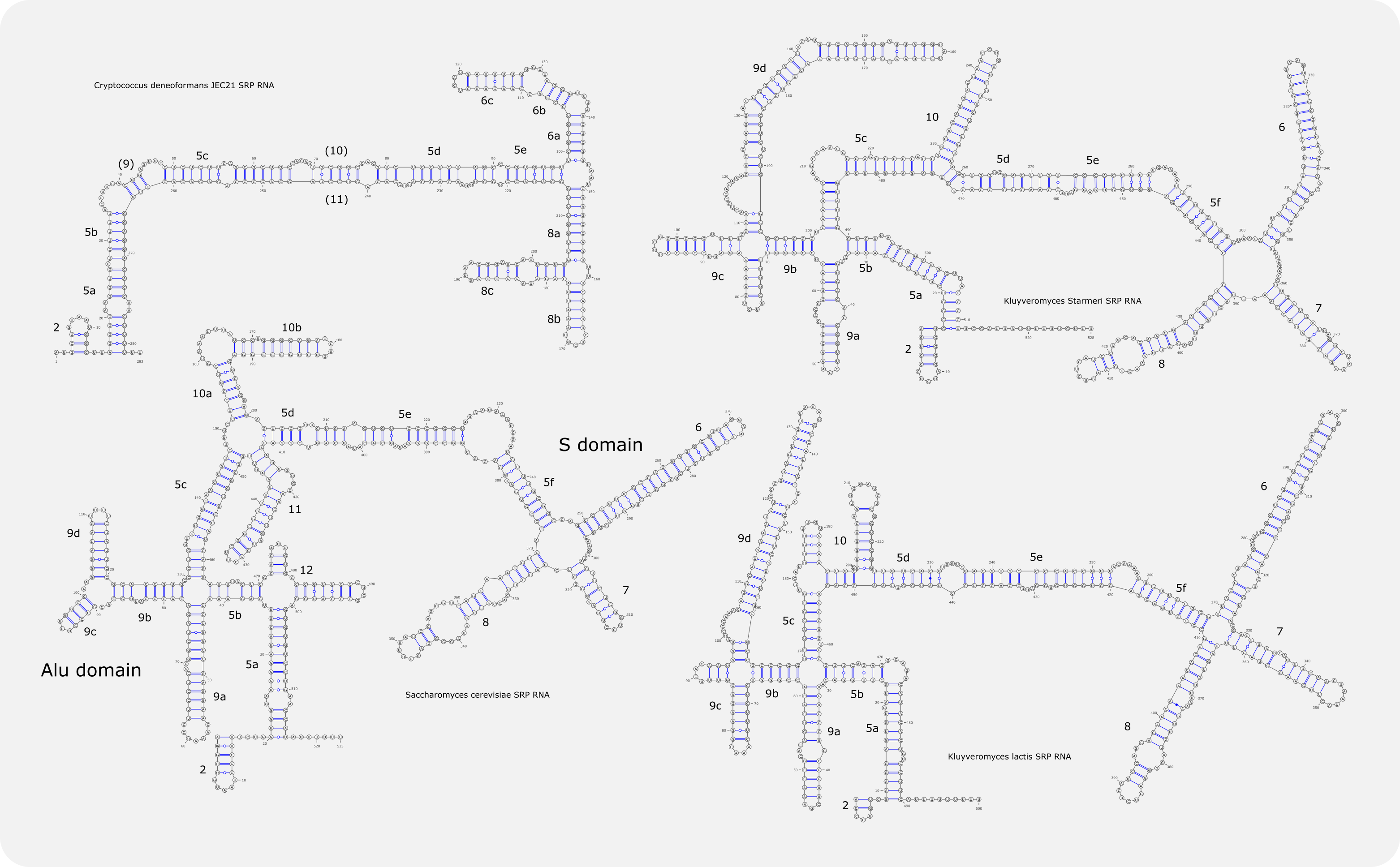

Models for fungal SRP RNA¶
Alignments of SRP RNAs from species related to Cryptococcus (top) or baker’s yeast (bottom) were made with JalView and the secondary structures (middle) with Varna. For the original and other models see [Rosenblad-2004, van.Nues-2007, Dumesic-2015] or below [1].
Compared to SRP RNA from bacteria, human or fungi like Candida, the RNA in baker’s yeast Saccharomyces cerevisiae and related species is marked by a very complex structure due to additional helices that protrude from the central rod or are part of an extended Alu domain [Rosenblad-2004, van.Nues-2004, van.Nues-2007]. These helices do not appear to be redundant and one helix harbours sequences that, on the level of DNA, form part of the RNA polymerase III promoter. Close inspection of some SRP RNAs suggest that the ensemble of structural elements found to be involved in SRP function are not as constrained as the literature suggests. For example, helix 8, which contains internal bulges essential for Srp54 association, appears to have an asymmetric bulge expanded into a helical segment (8b) in Cryptococcus [Dumesic-2015], and, surprisingly, the highly conserved GUAAU element in the Alu-domain linked to SRP14 function was not found at the 5’ end of Kluyveromyces SRP RNAs [2].
>AE017341.1:c775647-775365 Cryptococcus neoformans var. neoformans JEC21 chromosome 1, complete sequence ATGCTGTAATGGCTTAGGGGAAGGTGCTCTTTTACTTTCACGGAGTTCCTCTCTTCAGCGCTGCCACATT GCGGCCACGTCTCCTCTAGCCCGTGTGTTCGAGTCCCACCAAGATCTCATGATTTTGTGTTTGGGTTTAA CTCGGTAATCAAGCCAGTTTTCATTTCTCGTAAATGACAGATAGGCCGGTAACGGATCATTCTGGCTGGT TTGGCATACGTGCTTTAAGAGGATTGGACAGTCGTAGCAGCGCAGAGAGAGCCGGGAGGACACCAACTTT TTT
Notes¶
>JEC21_SRP_RNA AE017341.1:c775647-775365 Cryptococcus neoformans var. neoformans JEC21 chromosome 1, complete sequence
AUGCUGUAAUGGCUUAGGGGAAGGUGCUCUUUUACUUUCACGGAGUUCCUCUCUUCAGCGCUGCCACAUUGCGGCCACGUCUCCUCUAGCCCGUGUGUUCGAGUCCCACCAAGAUCUCAUGAUUUUGUGUUUGGGUUUAACUCGGUAAUCAAGCCAGUUUUCAUUUCUCGUAAAUGACAGAUAGGCCGGUAACGGAUCAUUCUGGCUGGUUUGGCAUACGUGCUUUAAGAGGAUUGGACAGUCGUAGCAGCGCAGAGAGAGCCGGGAGGACACCAACUUUUUU
..(((.....)))..(((((..((((.((((((.......(((.....(((((((..(((((((.....((((((...((((((((((((.(((((((((((((((((.(((((((....)))))))....))))....))))))....(((((((((..((((((.....))))))((((..(((((....))).))..)))))))))))))))))))).)))...))))))...))).))))))))))))).))))))))))))))))))))..)))))..
::cccnnnnnccc::EEEEEKKDDDD{FFFFFFPPPPPPPCCCNNNNNGGGGGGGKKGGGGGGGNNNNNFFFFFFLLLCCCFFFFFFCCC{GGGGGGGffffffdddd[gggggggmmmmggggggg4444dddd4444ffffff::::IIIIIIIII::ffffffnnnnnffffffdddd!!bbcccmmmmccc]bb22ddddIIIIIIIIIGGGGGGG}CCCZZZFFFFFFZZZCCC}FFFFFFGGGGGGG}GGGGGGGCCCFFFFFFDDDDYYEEEEE::
>SacCer_CP092957 CP092957.1:c144246-143724 Saccharomyces cerevisiae S288C chromosome V
AGGCUGUAAUGGCUUUCUGGUGGGAUGGGAUACGUUGAGAAUUCUGGCCGAGGAACAAAUCCUUCCUCGCGGCUAGACACGGAUUGCACGCCCUUUGGGCAAGGGAUAGUUCUCUAUUCCGCACCGUGCCCUGUUGUGGCAACCGUCUUUCCUCCGUCGUAAAUUUGUCCUGGGCAGAGCUGUCUGCCCGGAGGCGGGAGAGUCCGUUCUGAAGUGUCCCGGCUAUAAUAAAUCGAUCUUUGCGGGCAGCCCGUUGGCAGGAGGCGUGAGGAAUCCGUCUCUCUGUCUGGUGCGGCAAGGUAGUUCUGGGUCCUUAGGGGCUCCACCUUCACCGCUGUUAGGGGAGUUUUAUCCAGCGUCAGCAAAGGUGACCCGUGAUGGAGGCGGCCGGGAUAGCACAUAUCAGUCGGAUAAUUGUGCAAGUUGAUCGCUUCGGCGGUUUAAUUUGGCGGUGCCAUCAGGAUUUACUCGCACAUUGUGGCCGUUCCCUCGGGGAUGGAGUGUGUCCUGAACCAUAUUUUUU
(((((.....)))))....((((..((((((((((.(((((((((((((((((((.......))))))..)))))))(((((..(((..((((...))))..(((((((....))))))))))))))).((((..(((((.((((((....(((((((.........(((.(((((((....))))))))))))))).)).(((((..((((.(((((((((((............((((((((((..((((((((((((((((((.((....)))))))).))))).)))).))).....((.(((((....))).)).)).((((((....((((.....(((......))).....)))).)))))).))))))).)))...)))))))...))))...)))).)))))((((....((((((.(((...))))))))))).))))))))))))))))))))..))).(((...))).(((((((....))))))).))))))))))..)))).......
eeeeennnnneeeee::::DDDDKKJJJJJJJJJJ:CCCCCCgggggggffffffpppppppffffff22gggggggEEEEEKKCCC::ddddllldddd::gggggggmmmmgggggggCCCEEEEE:DDDDKKEEEEE{FFFFFF::::bbeeeee(((((((((ccc[gggggggmmmmgggggggccceeeee]bb:EEEEEKKDDDD{DDDDGGGGGGGUUUUUUUUUUUUCCCGGGGGGG::cccddddeeeeeffffff[bbmmmmbbffffff]eeeee]dddd]ccc:::::bb[bbcccmmmmccc]bb]bb:ffffff####dddd$$$$$cccooooooccc55555dddd]ffffff:GGGGGGG}CCCZZZGGGGGGGZZZDDDDZZZDDDD}EEEEEbbbb####ffffff[ccclllcccffffffbb]bbFFFFFFEEEEEDDDDCCCYYCCC:ccclllccc:gggggggmmmmggggggg:JJJJJJJJJJYYDDDD:::::::
>KluLac_CP042456 CP042456.1:1273402-1273901 Kluyveromyces lactis strain CBS 2105 chromosome 2
ACCCUGUCGAAGGAGUGUGAUGCUUGGACAUGUGGCUCCGGUUCGAAACCGCACAGUCACACCGGGUCUCUUUCAAACGGAAGAGGUUUCACAAUGGUAUCCCAAACAGGCAUGAAAUGUCCCGUAGUGGUUAUAUGCCAUUACCCGCUUUUCGUGUCUGGCCGCCCGGUCUACGCGUUCAUUUGUUUGUGUAACUGCUUUGGACAGUUGGAUCUCGGUCCGGUUGUCACGUGGUCGACGUGCCCCACCGUUACAAAAUUGUUGCGGGUUACCCUUUGUGGUGGCUCGGUCGCAUGAGAAGUCUGCGGUUCGAAGCCCAACUGGUAACGAUUUAGCAUUUCAACUAAUACUUGAAGCUAGUUUAGUGUUUAACGUUGUAGGGGAGUCUCAUCCAGCACAACAAACACUGCCCGUGACGAGGUGGUGUGGAUUGCACGUCCUAUGGUUAAGCAGUGCGUGUGGGUUCAGAGGCAGACAUACACUGUCCUUCAUUUUUUUUU
((...)).((((((((((.((((((((((.(((((((.(((((...)))))...)))))))(((((((((((((....)))))))(((....)))(((........(((((((((((.((...((((((((.....))))))))..)).))))))))))))))))))))((((((((.....((((....))))((((.((((((.(........).))))))((((((((.....((((((((((((((((......(((((((((((((((..(((....((((((((((((.((....)))))))).))).))))))..))))).((.((((..((((((.......))))))))))))(((((((...((((((...(((......)))...)))))))))))))))))))))))))))))).))...))))))).))))).))))))))))))).)))))).))).....))))))))..)))))..........
bblllbb:EEEEEEEEEE{CCCCCCDDDD:ggggggg[eeeeellleeeee333gggggggFFFFFFgggggggmmmmgggggggcccmmmmcccccc''''''''===========[bb"""hhhhhhhhnnnnnhhhhhhhh22bb]===========cccFFFFFFBBFFFFFF:::::ddddmmmmddddDDDD:ffffff[aqqqqqqqqa]ffffffCCCEEEEENNNNNGGGGGGGBBGGGGGGGOOOOOOJJJJJJJJJJeeeee!!ccc####ccccccffffff[bbmmmmbbffffff]ccc]cccccc22eeeee:bb[dddd!!ffffffpppppppffffffddddbbggggggg"""ffffff"""cccooooooccc333ffffffgggggggJJJJJJJJJJGGGGGGG}BBZZZGGGGGGG}EEEEE}CCCDDDDFFFFFF}BBDDDD}CCC?????CCCEEEEEYYEEEEE::::::::::
>KluMar_AP014601 AP014601.1:c280853-280319 Kluyveromyces marxianus DNA, chromosome 3, complete genome, strain: NBRC 1777
AAUUGGCCUUUCUGGAAGUCAGAGAUGUGAUACUUGGAUAAGUGGCUCCGGUUCGAAACCGCACUGUCACUCCGGGUCCCUUGUAUUAGGGGUCUCCACGGUACUAUUAAACAGCAUCGGUAGUCUCUUGUGAGACCCGUCCUACUCGUAGGCGGGUAACAAGCUGCUGGUGUUGGCCGCCCGGCCCAGACGUCCCACUAGAACUUCUUCCGAUCUGAGAGAUUUUAUCGUCUCACAGGUCACGUGGUCGACUUGUCCCACCGUUUCAAAAUUGUUGCGGGUCUUUCCUGUAUGGUGGCGCGACUGCGCGAGAAAUCCGCGGUUCGAGUCCAACUGGUAACGAUUUAGCAAAUUUUCCCAUCUCGCAUGGGAAAGCCCGCUAAUUAGUGUUUACUGUUGAAGGGGAGUCUCAUCCAGCACAACGAACACUGCCCGUGACGAGGUGGUGUGGAUUGCAAGUCCCAUGUAUAAGGAGUACGUUUGGAUCCAGAUGUAUCAGCUAGCCUGACCAAUUUUUUU
((((((.(((.....)))((((.....((((((.(((((.((((((..(((((...)))))....))))))((((((((((((...)))))).......(((.........((((((((((((...(((((...(((((.((((....))))))))).)))))))))))))))))))))))))).((((((((.........(((((((..((((((.(((((......))))).))))))((((((..((((((((((((((((......((((((((((((.((((.((....(((((.(((((((.((....)))))))))))...))))).)).)).)).(((((...(((((((((.....)))))))))...))))).(((((((...(((((....(((......)))....))))))))))))))))))))))))))))).))...)))))))))))))..))))))))))))))))))))...))))))......)))))))))).....
FFFFFF:cccnnnnncccDDDDNNNNNFFFFFF{EEEEE:ffffff!!eeeeellleeeee4444ffffffFFFFFFfffffflllffffff:::::::ccc(((((((((============"""eeeee"""eeeee[ddddmmmmddddeeeee]eeeee============cccFFFFFF:HHHHHHHHRRRRRRRRRGGGGGGG::ffffff[eeeeeooooooeeeee]ffffffFFFFFFKKGGGGGGGBBGGGGGGGOOOOOOJJJJJJJJJJbb[bbbb[bb####cccbb[ggggggg[bbmmmmbbgggggggbb333cccbb]bb]bb]bb:eeeee"""iiiiiiiiinnnnniiiiiiiii333eeeee:ggggggg"""eeeee####cccooooooccc4444eeeeegggggggJJJJJJJJJJGGGGGGG}BBZZZGGGGGGGFFFFFFYYGGGGGGGHHHHHHHHEEEEEZZZFFFFFF______DDDDFFFFFF:::::
>KluSta WACN01000003.1:746854-747381 Kluyveromyces starmeri strain UFMG-CM-Y3682 NODE_3_length_972539_cov_72.8531, whole genome shotgun sequence
ACUCCCUCUAGGGGUGCGGUCUGUCUGCAGACAUGUGACACCGGUUCGAAACCGCACGGUCACUCCGGGUCUUUCUUCUGAAAGAGAUUCCGCCUUCGGGCGGUUUUCGGUACUCCCAUAAACAGCGAGGACGGGGCAGUUCUUGUCACGUGAUUAGGUAUCCUAGCCACGUGACACUGUCUCCCUCUCUGGCCGCUCGGUCAAUGCAAGCUAACGGGAGCCUUCAUCGGCACAGGACGAACCUGUUCUUUCUGUGUCACGUGGUGGACUUGUCCCACCGUUUGAAAAUUGUUGCGGGUAACAUUUGGUGGUUUGGCCGCGCGAGAAGUCCGCGGUUCGAAGCCCAUCUGGUAACGACCUUGGUAUUUAGUCUAAUGACGAUACCAAACCAGUGUUUAACGUUGAAGGGGAGUCUCAUCCAGCACAACGAACACUGCUCGUGACGAGGUGGUGAGGAUUGCAAGUCCCAUGGUGAGAAGGUCCGCAUUGGUCUUAGCAGAUGGUUAGCCGUUCGAUUAUUUUUUGUUU
(((((.....)))))(((((((((((((((((..(((((..(((((...)))))....))))).(((((((((((....)))))(((..(((((....))))).))).(((...........(((.((((..((((((((....((((((((.(((((...))))).)))))))))))))))))))).)))))))))))).((((((.........(((.(((((.(((((((((((.(((....))).)))))))))((((((..((((((((((((((((......((((((((((....(((.(((((((((((((((.((....))))))))).)))..))))).))).......((((((((..(((....)))))))))))..(((((((...(((((....(((......)))....))))))))))))))))))))))))))))).))...))))))))))).))))))))))))))))))))))..))))))))...))))).................
eeeeennnnneeeeeEEEEEHHHHHHHHDDDD::eeeee!!eeeeellleeeee4444eeeee:FFFFFFeeeeemmmmeeeeeccc!!eeeeemmmmeeeee]ccc:ccc+++++++++++ccc[dddd!!hhhhhhhh####hhhhhhhh[eeeeellleeeee]hhhhhhhhhhhhhhhhdddd]ccccccFFFFFF:FFFFFFRRRRRRRRRCCC{EEEEE{BBiiiiiiiii[cccmmmmccc]iiiiiiiiiBBDDDDKKGGGGGGGBBGGGGGGGOOOOOOJJJJJJJJJJ::::ccc[eeeeecccggggggg[bbmmmmbbggggggg]ccc22eeeee]ccc:::::::hhhhhhhh!!cccmmmmccchhhhhhhh::ggggggg"""eeeee####cccooooooccc4444eeeeegggggggJJJJJJJJJJGGGGGGG}BBZZZGGGGGGGDDDD}BBBBEEEEECCCFFFFFFDDDDYYHHHHHHHHZZZEEEEE:::::::::::::::::
For example blasting at NCBI the SRA archive for TSS RNA-seq libraries of Kluyveromyces Marxianus (like SRX905361, found by https://www.ncbi.nlm.nih.gov/sra/?term=Kluyveromyces+TSS) with AAATCCCGATTTTCACAATTGGCCTTTCTGGAAGTCAGAGATGT, expected to cover the 5’ end of SRP RNA, finds as nearest match AATTGGCC… suggesting this as its 5’ end. Checking upstream regions of various Kluyveromyces SRP loci did not suggest the presence of an intron or an obvious candidate that would resemble the GUAAU element, only a CACGTGA/C sequence was shared between the assessed Kluyveromyces loci (CP042456.1:1273282-1273901 Kluyveromyces lactis strain CBS 2105 chromosome 2, AP014601.1:c280993-280319 Kluyveromyces marxianus DNA, chromosome 3, complete genome, strain: NBRC 1777, JAGEVD010000013.1:c72707-71920 Kluyveromyces sp. PCH397 tig00000021, whole genome shotgun sequence, CP015056.1:c305047--304369 Kluyveromyces marxianus strain FIM1 chromosome 3, partial sequence, LDJA01000001.1:1278725-1279416 Kluyveromyces marxianus strain IIPE453 Scaffold_1, whole genome shotgun sequence, CCBQ010000045.1:1141109-1141801 Kluyveromyces dobzhanskii CBS 2104 WGS project CCBQ000000000 data, contig 00015, whole genome shotgun sequence, WACN01000003.1:746708-747381 Kluyveromyces starmeri strain UFMG-CM-Y3682 NODE_3_length_972539_cov_72.8531, whole genome shotgun sequence, JAKVQO010000004.1:1023455-1024153 Kluyveromyces sp. yHMH660 NODE_4_length_1110509_cov_22.247_ID_13521, whole genome shotgun sequence, JAKVQO010000004.1:1023455-1024153 Kluyveromyces sp. yHMH660 NODE_4_length_1110509_cov_22.247_ID_13521, whole genome shotgun sequence, AEAV01000057.1:40618-41237 Kluyveromyces wickerhamii UCD 54-210 contig00060, whole genome shotgun sequence, QYLQ01000002.1:867685-868285 Kluyveromyces nonfermentans strain NRRL Y-27343 flattened_line_2, whole genome shotgun sequence.
The program [Mattei-2014] was run with the command java -jar BEAR_encoder.jar -i test.fasta -o out, in which test.fasta contained the first three lines describing name, sequence and fold (ensure any - is replaced by . in the dot-bracket line); out would be empty if the input was not correct; otherwise it would have the four lines shown.
Image source Tremellomycetes’ SRP RNA:
# STOCKHOLM 1.0
#=GS JEC21/1-283 DE SRP-RNA
JEC21/1-283 ATGCTGTAATGGCT-TAGGGGAAGGTGCTCTTTT--ACTTTCACGGAGTTCCTCTCTTCAGCGC-TGC-C-ACATTGCGGC--CAC---GTCTCCTCT-AGCCCGTGTGT-TCGAG-T---CCCACCAAGATCTCATGATTTTG-----------TGTT--------------TGGG-----TTTA---------A-CTCGG-TAATCAA-GCCAGT-----------TT-------TCATTT------CTCGT------AAATGA------------CAGATAGGCCGGTAACGGATCATTCTG-GCTGG-TTTGGCATACGTGCTTTAAGAGGATTGGACAGTCGTA-GCA---GCGCAGAGAGAG-CCGGGAGGACACCAACTTTTTT--
#=GS XL280/1-283 DE AWPH01000001.1:c775657-775328 Cryptococcus neoformans var. neoformans chromosome 1 contig001, whole genome shotgun sequence
XL280/1-283 ATGCTGTAATGGCT-TAGGGGAAGGTGCTCTTTT--ACTTTCACGGAGTTCCTCTCTTCAGCGC-TGC-C-ACATTGCGGC--CAC---GTCTCCTCT-AGCCCGTGTGT-TCGAG-T---CCCACCAAGATCTCATGATTTTG-----------TGTT--------------TGGG-----TTTA---------A-CTCGG-TAATCAA-GCCAGT-----------TT-------TCATTT------CTCGT------AAATGA------------CAGATAGGCCGGTAACGGATCATTCTG-GCTGG-TTTGGCATACGTGCTTTAAGAGGATTGGACAGTCGTA-GCA---GCGCAGAGAGAG-CCGGGAGGACACCAACTTTTTT--
#=GS H99/1-282 DE NPNB01000014.1:c801482-801153 Cryptococcus neoformans var. grubii strain H99 Chr_1, whole genome shotgun sequence
H99/1-282 ATGCTGTAATGGCT-TAGGGGAAGGTGCTCTTTT--ACTTTCACGGAGTTCCTCTCTTCAGCGC-TGC-C-ACATTGCGGC--CAC---GTCTCCTCT-AGCCTGTGTGT-TCGAG-T---CCCACCAAGATCTCATGATTTTG-----------TCTT--------------TGGG-----TTT----------A-CTCGG-TAATCAA-GCCAGT-----------TT-------TCATTT------CTCGT------AAATGA------------CAGATAGGCCGGTAACGGATCATTCTG-GCTGG-TTTGGCATACATGCTTCAAGAGGATTGGACAGTCGTA-GCA---GGGCAGAGAGAG-CCGGGAGGACACCAACTTTTTT--
#=GS EJB2/1-281 DE ATAL01000262.1:c19201-18903 Cryptococcus gattii EJB2 cont1.262, whole genome shotgun sequence
EJB2/1-281 ATGCTGTAATGGCT-TAGGGGAAGGTGCTCTTTT--ACTTTCACGGAGTTCCTCTCTTCAGCGC-TGC-C-ACATTGCGGC--CGC---GTCTCCTCT-AGCCCGTGTGT-TCGAG-T---CCCACCAAGATCTCATGATTTTG-----------TCTT--------------TGGG-----TTT----------A-CTCGG-TAATCAA-GCCAGT-----------TC-------TCATTT------CTCGT------AAATGA------------CAGATAGGCCGGTAACGGATCATTCTG-GCTGG-TTTGGCATACATGCTTCAAGAGGATTGGACAGTCGTA-GCA---GCGCAGAGAGAG-CCGGGAGGACACCAACTTTTT---
#=GS IND107/1-281 DE ATAM01000015.1:c321711-321413 Cryptococcus gattii VGIV IND107 cont2.15, whole genome shotgun sequence
IND107/1-281 ATGCTGTAGTGGCT-TAGGGGAAGGTGCTCTTTT--ACTTTCACGGAGTTCCTCTCTTCAGCGC-TGC-C-ACATTGCGGC--CGC---GTCTCCTCT-AGCCCGTGTGT-TCGAG-T---CCCACCAAGATCTCATGATTTTG-----------TCTT--------------TGGG-----TTT----------A-CTCGG-TAATCAA-GCCAGT-----------TC-------TCATTT------CTCGT------AAATGA------------CAGATAGGCCGGTGACGGATCATTCTG-GCTGG-TTTGGCATACATGCTTCAAGAGGATTGGACAGTCGTA-GCA---GCGCAGAGAGAG-CCGGGAGGACACCGACTTTTT---
#=GS CA1280/1-281 DE ASCN01000003.1:c94335-94039 Cryptococcus gattii CA1280 cont1.3, whole genome shotgun sequence
CA1280/1-281 ATGCTGTAATGGCT-TAGGGGAAGGTGCTCTTTT--ACTTTCACGGAGTTCCTCTCTTGAGCGC-TGC-C-ACATTGCGGC--CGC---GTCTCCTCT-AGCCCGTGTGT-TCGAG-T---CCCACCAAGATCTCATGATTTTG-----------TCTT--------------TGGG-----TTT----------A-CTCGG-TAATCAA-GCCAGT-----------TC-------TCATTT------CTCGT------AAATGA------------CAGATAGGCCGGTAACGGATCATTCTG-GCTGG-TTTGGCATACGTGCTTCAAGAGGATTGGACAGTCGTA-GCA---GCGCAGAGAGAG-CCGGGAGGACACCAACTTTTT---
#=GS R265/1-281 DE NPNA01000007.1:c755739-755432 Cryptococcus gattii VGII strain R265 tig00000008, whole genome shotgun sequence
R265/1-281 ATGCTGTAATGGCT-TAGGGGAAGGTGCCCTTTT--ATTTTCACGGAGTTCCTCTCTTCAGCGC-TGC-C-ACATTGCGGC--CGC---GTCTCCTCT-AGCCCGTGTGT-TCGAG-T---CCCACCAAGATCTCATGATTTTG-----------TCTT--------------TGGG-----TTT----------A-CTCGG-TAATCAA-GCCAGT-----------TC-------TCATTT------CTCGT------AAATGA------------CAGATAGGCCGGTAACGGATCATTCTG-GCTGG-TTTGGCATACATGCTTCAAGGGGATTGGACAGTCGTA-GCA---GCGCAGAGAGAG-CCGGGAGGACACCAACTTTTT---
#=GS Ram5/1-281 DE ASCM01000040.1:40597-40904 Cryptococcus gattii VGII Ram5 cont1.40, whole genome shotgun sequence
Ram5/1-281 ATGCTGTAATGGCT-TAGGGGAAGGTGCCCTTTT--ATTTTCACGGAGTTCCTCTCTTCAGCGC-TGC-C-ACATTGCGGC--CGC---GTCTCCTCT-AGCCCGTGTGT-TCGAG-T---CCCACCAAGATCTCATGATTTTG-----------TCTT--------------TGGG-----TTT----------A-CTCGG-TAATCAA-GCCAGT-----------TC-------TCATTT------CTCGT------AAATGA------------CAGATAGGCCGGTAACGGATCATTCTG-GCTGG-TTTGGCATACATGCTTCAAGGGGATTGGACAGTCGTA-GCA---GCGCAGAGAGAG-CCGGGAGGACACCAACTTTTT---
#=GS CryDep/1-283 DE AWGK01000011.1:154376-154669 Cryptococcus depauperatus CBS 7841 supercont1.11, whole genome shotgun sequence
CryDep/1-283 ATGCTGTCATGGCT-TATGGGAAGGTGTGCTTTT--ATTCTCACGGAGTTCCTCTCTTCAGAGC-TCC-T-CTATTGCGGC---GC---GTCTCCTCTTAGCTCGTGTGT-TCGAG-T---CCCACCAAAGTCTCATGATTTTG-----------TGTT--------------TGGG-----TTT----------A-CTCGG-TAATCAA-GTCAGT-----T----TTA--------CATTTC-----CTTG------GAAATG---------G---CAGATAGGCCGGTAACGGATCATTCTG-GCTGA-CTTGGCATTCGAGCTTCAAGAGGATTGGACAGTCGTA-GGA---GCGCAGAGAGAG-CCGGGAGTATACCAACTTTTT---
#=GS CryDep-2/1-283 DE AWGL01000006.1:184646-184937 Cryptococcus depauperatus CBS 7855 supercont2.6, whole genome shotgun sequence
CryDep-2/1-283 ATGCTGTCATGGCT-TATGGGAAGGTGTGCTTTT--ATTATCACGGAGTTCCTCTCTTCAGAGC-TCC-T-CTATTGCGGC---GC---GTCTCCTCTTAGCTCGTGTGT-TCGAG-T---CCCACCAAAGTCTCATGATTTTG-----------TGTT--------------TGGG-----TTT----------A-CTCGG-TAATCAA-ATCAGT-----T----TTA--------CATTTC-----CTTG------GAAATG---------G---CAGATAGGCCGGTAACGGATCATTCTG-GCTGA-CTTGGCATTCGAGCTTCAAGAGGATTGGACAGTCGTA-GGA---GCGCAGAGAGAG-CCGGGAGTATACCAACTTTTT---
#=GS CryWin/1-287 DE AWGH01000015.1:213774-214072 Cryptococcus wingfieldii CBS 7118 supercont1.15, whole genome shotgun sequence
CryWin/1-287 ATGCTGTTATGGCT-TAAGGGAAGATGCTCTTCT--ACTATCACGGAACTCCTCTCTTCAGCGC-TCC-C-ATAATACAAC--CTC---GTCTCCTCT-AGCGTGTGTGT-TCGAG-C---CCCACCAATGTCTCATGATTTTG----------CGTTT--------------TGGG----TTTAAA--------G-CTCGG-TAATCAA-ATCAGT-----T----CTA--------CATTTC-----TTT-------GAAATG---------G---CAGATAGGCCGGCAACGGATCATTCTG-GCTGG-TTTGGCATATGCGCTTCAAGAGGATTGGACAGTTGTA-GGA---GCGCAGAGAGAG-CCGGGAGAGTATCAACTTTTTT--
#=GS CryFlo/1-286 DE RRZH01000013.1:778013-778306 Cryptococcus floricola strain DSM 27421 chromosome 12, whole genome shotgun sequence
CryFlo/1-286 ATGCTGTTATGGCT-TAAGGGAAGATGCTCTTTT--ATTATCACGGAACTCCTCTCTTCAGCGC-TCC-C-ATAATACAAC--CTC---GTCTCCTCT-AGCGTGTGTGT-TCGAG-C---CCCACCAATGTCTCATGATTTTG----------CGTTT--------------TGGG----TTTAAA--------G-CTCGG-TAATCAA-ATCAGT-----T----CTA--------CATTTC-----TTT-------GAAATG---------G---CAGATAGGCCGGCAACGGATCATTCTG-GCTGG-TTTGGCATATGCGCTTCAAGAGGATTGGACAGTTGTA-GGA---GCGCAGAGAGAG-CCGGGAGAGTATCAACTTTTT---
#=GS CryAmy/1-286 DE MEKH01000003.1:c440117-439822 Cryptococcus amylolentus CBS 6273 supercont2.3, whole genome shotgun sequence
CryAmy/1-286 ATGCTGTTATGGCT-TAAGGGAAGATGCTCTTTT--ATTATCACGGGACTCCTCTCTTCAGCGC-TCC-C-ATAATACAAC--CTC---GTCTCCTCT-AGCGTGTGTGT-TCGAG-C---CCCACCAATGTCTCATGATTTTG----------CGTTT--------------TGGG----TCTAAA--------G-CTCGG-TAATCAA-ATCAGT-----T----CTA--------CATTTC-----TTT-------GAAATG---------G---CAGATAGGCCGGCAACGGATCATTCTG-GCTGG-TTTGGCATATGCGCTTCAAGAGGATTGGACAGTTGTA-GGA---GCGCAGAGAGAG-CCGGGAGAGTATCAACTTTTT---
#=GS CrySki/1-285 DE BCHT01000012.1:c526360-526070 Cryptococcus skinneri DNA, scaffold: scaffold_11, strain: JCM 9039, whole genome shotgun sequence
CrySki/1-285 CAGCTGTAGCGGCT-TTTGGGAAGGTG-TCTATT-TTCATTCACGGAGTTCCTCTCCTCAGCGC-GCC-C-TCGTTACAACC--CC---GTCTCCTCT-AGCACGAGTGT-TCGAGCC----CCAGCGAAATCTCATGATTTTG-----------AT---------------TTGG------TCATT--------GTCTCGG-TGATCAA-GCCAGT-----T-----TC-----CCACATT-------CTCG--------AATGTGG----------CGGATAGGCCGGTAACGGATCAGTCCG-GCTGG-TTTGGCATTTGGGCTTAAAGAGGTTGGGACGGTTGTA-GGA---GCGCAGGGAGAG-CCGGGTGGGCACCAACTTTTTT--
#=GS CryFagi/1-301 DE BCHU01000002.1:c886806-886496 Cryptococcus fagi DNA, scaffold: scaffold_1, strain: JCM 13614, whole genome shotgun sequence
CryFagi/1-301 ACGCTGTAATGGCT-TTTGGGAAGGTG-TTCATT-TTATTTCACGGAATTCCTCTCCTCAGAGC-TCC-C-TCGTTACAACC--TT---GTCTCCTCT-AGCACGAGTGT-TCGAGCC----CCATCAGGATCTCATGATTCTG-T-AGAAT---ATCT-ATTCT-TTAA--ATGG------TTCC---------GTCTCGG-TGATCAA-GCTAGT-----T------------TTACATATA-----CTT-------TGTATGTGG----------CAGATAGGCCGGTAACGGATCAGTCTG-ACTTG-TTTGGCATTTGAGCTTAAAGAGGTTAGGACGGTTGTA-GGA---GCGTAGGGAGAG-CCGGATGGGCACCAACTTTTT---
#=GS CryVH57/1-288 DE JANBXI010000024.1:c27124-26828 Cryptococcus sp. VH57_BW 24, whole genome shotgun sequence
CryVH57/1-288 ATGCTGTAATGGCTTATTGGGAAGGTG-TTCTTT-TTCACTCACGGAGTTCCTCTCTCCAGCGC-TCC-C-ATATTGCGTC--ATT---GTTTCCTCT-AGCCCCAGTGT-TCGAG-T---CCCAGCAAAGTCTCATGATTTTG-----------TATA--------------TGGG---TCTCAAT--------A-CTCGG-TAATCAA-GTTAGT-----T----CTA--------CATTTC-----ATTA------GAAATG---------G---CAGATAGGCCGGCAACGGATCATTCTG-GCTAA-TTTGGCATTGGTGCTTCAAGAGGTTTGGACAGGCGTA-GGA---GCGCAGGGAGAG-CCGGGAGGGCACCAACTTTTT---
#=GS KocImp/1-292 DE NBSH01000007.1:85321-85638 Kockovaella imperatae strain NRRL Y-17943 BD324scaffold_7, whole genome shotgun sequence
KocImp/1-292 TCGCCGTCGTGGCT-CTTGGGAAGGTG-GTCATT-TTCTTTCACGGAACACCTCTCTTCAGCGC-GCC-CCTTTGTACGAA--TATC--GTCTCCTCT-AGCATCAGTGT-TCGAG-T---CTCACCGAAGTCTCATGACTTTG--------TGAACTACTT-----------TGAG-----TTTTA--------A-CTCGG-TAATCATC-TCAGT-----------AT-------ACATCT-----CTCTGC------AGATGT------------CAGATAGGCCGGTTACGGATCATTCTG-GCTGA-TTTGGCATTGTTGCTCCAAGAGGTTTGGACTGTCGTA-GGATTGTCGCAGGGAGAG-CCGGGTGATCACCAACTC------
#=GS BulAlb/1-284 DE CAMYTR010000137.1:253624-253915 Bullera alba genome assembly, contig: jcf7180000012238, whole genome shotgun sequence
BulAlb/1-284 TCGCTGTAGTGGCT-TTTGGGAAGATG-TCTGTT-TTTCATCACGGGTTTCCTCTCGTCAGCGC-TCC-C-TCGTTATGCTT--TA---GTCTCCTCT-AGCACGAGTGT-TCGAG-C---ACCACCCAGATCTCACGATTTGG----------TTGT---------------TGGT-----TCT----------G-CTCGG-TAATCAA-GTCAGT----TC----TTT-------ACTCC-------TTCT--------GGAGT--------GA--CAGATAGGCCGGCAACGGATCAATCTG-GCTGA-TTTGGCATTCGGGCTTCAAGAGGATTGGACATGTATA-GGA---GCGCAGCGAGTG-CCGGGTGGACATCAACCTTTTT--
#=GS VisVic/1-289 DE JADPYG010000092.1:c218279-217975 Vishniacozyma victoriae isolate T18_1_22C Contig_92, whole genome shotgun sequence
VisVic/1-289 ATGCTGTAATGGCTTACCGGGAAGATG-CTCATT-TATCATCACGGAATTCCACTCTTCAGCGC-TCC-C-TTTGTGCAGC--AAA---GTCTCCTCT-AGCATCAGTGT-TCGAG-C-C--CCACCCTTTCCTCACGGTTTGG----------TTGTT--------------TGG-----TTATAC--------G-CTCGG-TGATCAA-GTTAAT----------ATTT------TCAACC-----CTTTGT------GGTTGA------------CAGATAGGCCGGCAACGGATCATTCTG-GTTGA-TTTGGCATTGATTCTTCAAGAGGATAGGACGGTTGTT-GGA---GCGTAAAGAGTG-CCGGTTGGGTATCGACCTTTTT--
#=GS TetT11/1-289 DE JADPYF010000126.1:c218262-217963 Tetracladium sp. T11_2_10C Contig_126, whole genome shotgun sequence
TetT11/1-289 ATGCTGTAATGGCTTACCGGGAAGATG-CTCATT-TATCATCACGGAATTCCACTCTTCAGCGC-TCC-C-TTTGTGCAGC--AAA---GTCTCCTCT-AGCATCAGTGT-TCGAG-C-C--CCACCCTTTCCTCACGGTTTGG----------TTGTT--------------TGG-----TTATAC--------G-CTCGG-TGATCAA-GTTAAT----------ATTT------TCAACC-----CTTTGT------GGTTGA------------CAGATAGGCCGGCAACGGATCATTCTG-GTTGA-TTTGGCATTGATTCTTCAAGAGGATAGGACGGTTGTT-GGA---GCGTAAAGAGTG-CCGGTTGGGTATCGACCTTTTT--
#=GS TreFJI/1-302 DE JAKLMH010000005.1:273830-274145 Tremellales sp. FJII-L8-SW-PAB2 NODE_5_length_1036247_cov_87.416123, whole genome shotgun sequence
TreFJI/1-302 ACGCTGTAATGGCTTACTGGGAAGGTG-TGCATT-TCACATCACGGAGTTCCACTCATCCGCGC-TCC-C-ATTGTGCAAC-ATTAA--GTCTCCTCT-AGCACCAGTGT-TCGAG-C-C--CCAGCCCATCCTCAAGGATGTC---TTGG---TTGT---TTAA--AA----TGG------TTCC---------G-CTCGG-TGATCAA-GTCGAT----------ATAT------TCTACT----CTTTACTGT----AGTAGA------------CAGATAGGCCGGCAACGGATCATTCTG-GTCGA-TTTGGCATTGGCGCTTCAAGAGGATAGGACGGTTGTT-GGA---GCGTAGTGAGTG-CCGGGTGTGCACCAACTCTTTTT-
#=GS TreFuc/1-300 DE BRDD01000111.1:c218982-218662 Tremella fuciformis NBRC 9317 DNA, KCNB80TF.111, whole genome shotgun sequence
TreFuc/1-300 CCGCTGTAGCGGCT-TTCGGGAAGGTG-TTTATT-TGTCATCACGGATTCACCCTCGTCAGCGC-TCC-C-ACTGTGCAAT--GCA---GTCTCCTCT-AGCGCCAGTGT-TCGAG-C-C--CCACCGTGACCTCATGGCCACG---------TTGACTT-------------TGG------TTCCTATAC----G-CTCGG-TGATCAG-ATCAGT-----T-----TA--------CACTTTC---CTAATAC---GAAAGTG---------G---CAGATAGGCCGGCAACGGATCATTCTG-ACTGCGTCTGGCATTGGCGCTTCAAGAGGATAGGACGGTTGTT-GGA---GCGTAGCGATGGCCCGGGTGGGCACCGACTCTTTTTT
#=GS NaeEnc/1-300 DE MCFC01000118.1:c11368-11058 Naematelia encephala strain 68-887.2 BCR39scaffold_118, whole genome shotgun sequence
NaeEnc/1-300 CAGCTGTCATGGCT-TTTGGGAAGGTG-ATTACT-TTTCATCACGGAGTTCCCCTCCTCCGAGC-TCC-C-ACATTACGAGATTTTCA-GTCTCCTCT-AGCGCCAGTGT-TCGAG-T-C--CCACCCATGTCTCAAGACGTGG---------TTGATTT-------------TGG------CACAC--------G-CTCGG-TAATCAACGTCAGT-----T-----TA----CGTTTCCGTAC---CTCGT-----GTACGGG-CG------G---CGGACAGGCCGGCAACGGATCAGTCTG-GCTGA-TTTGGCATTGGTGCTTCAAGAGGATAGGACACTCGTA-GGA---GCGCAGGGAGGG-CCGGGTGGTCACCAACTTTTT---
#=GS KwoSha/1-307 DE NQVO01000004.1:c344412-344094 Kwoniella shandongensis strain CBS 12478 scaffold00004, whole genome shotgun sequence
KwoSha/1-307 ACGCTGTAATGGCT-TTTGGGAAGGTG-TTCATT-TTCACTCACGGAACTCCTCTCTTCAGCGC-TCC-A-TCATTACGTT-ATTA---GTCTCCTCT-AGCACCAGTGT-TCGAG-T-C--CCACCAAAGTCTCATGATTTTG---------TCGTTTA-------------TGG--TTGTCTCATGTAAT---A-CTCGG-TAATTAA-GCTCGT-----T----CTA--------CATTTACTC--TTCG---GAGAAAATG---------G---CGGTTAGGCCGGCAACGGATCAGACCG-GCGTTGCTTAGCGTTGATGCTACAAGAGGATAGGACAAACGTA-GGAT--GCGCAGGGAGAG-CCGGGTGAATACCAACTCTTTTT-
#=GS PapLau/1-291 DE JDSR01001044.1:23278-23579 Papiliotrema laurentii RY1 contig_1081, whole genome shotgun sequence
PapLau/1-291 ATGCTGTAACGGCT-TTTGGGAAGATG-TCCATT-TACTACAACGGAATCAATCTCATCAGCGC-TCC-C-TGATTACATTT--CA---GTCTCCTCT-AGCACGAGTGT-TCGAG-C-C--TCACCCTTGTCTCAAGACGAGG-----------TGT---------------TGA------CTTT-C-------G-CTCGG-TGATCAA-ACCAGT-----------AT-------GTAATTTGCT-GTAAAA--GGCAA-TTAT------------CAGATAGGCCGGCAACGGATCATTCTG-CCTGGGGTTGGCATTTGGGCTTCAAGAGGATAGGACGGGTGTA-GGA---GCGCAGTGAGAG-CCGGGTGGTCATCAACCTTTTT--
#=GS KwoMan/1-316 DE ASQF01000024.1:c596838-596513 Kwoniella mangroviensis CBS 8886 cont1.24, whole genome shotgun sequence
KwoMan/1-316 ACGCTGTAATGGCT-TTTGGGAAGGTG-TTTATT-TATCCTCACGGAACTACTCTCTTTAGCGC-TCC-A-TCATTACGTTT-CAAA--GTCTCCTCT-AGCATCAGTGT-TCGAG-T-C--CCACCAAAGTCTCATGATTTTG---TGTCCC-TTCAC-GGGGTGT-TA---TGG------CTCTT--------A-CTCGG-TAATCAA-GTTAGT-T-CTA----CAAA-------TCACCTC----CAAAA----GAGGTGG----CC---TGG-CGAATAGGCCGGTAACGGATCAATTCG-ACTAATCTTGGCATTGATACTACCAGAGGATAGGACAGACGTA-GGTTA-GCGCAGGGAGAG-CCGGGTGAACACCAACTTTTT---
#=GS KwoBes/1-306 DE ASCK01000019.1:c33468-33134 Kwoniella bestiolae CBS 10118 cont1.18, whole genome shotgun sequence
KwoBes/1-306 ACGCTGTAATGGCT-TTCGGGAAGGTG-TTTATT-TATCCTCACGGAACTCCTCTCCTTAGCGC-TCC-A-TCATTACGTT-ATATAT-GTCTCCTCT-AGCATCAGTGT-TCGAG-T---GCCACCAAAGTCTCATGACTTTG--------CGTATTTA-------------TGGC-----TCTT---------A-CTCGG-TAATCAA-GTTAGT----------TTTA-----CAGATGATCCT--CAAAA--GGGATTCGTCTG------G---CGAATAGGCCGGTGACGGATCAATTCG-ACTAATCTTGGCATTGATACTACCAGAGGATTGGACGGACGTA-GGTTA-GCGCAGGGAGAG-CCGGGTGAACACCAACTTTTT---
#=GS KwoHea/1-305 DE ASQC01000111.1:100400-100729 Kwoniella heveanensis CBS 569 cont2.111, whole genome shotgun sequence
KwoHea/1-305 ACGCTGTAATGGCT-TTTGGGAAGGTG-TTGATT-TACTCTTACGGACCTCCTCTCTTCAGCGC-TCC-A-TCATTACGTA-CCTAT--GTCTCCTCT-AGCATCAGTGT-TCGAG-T-C--CCAGCAGAGTCTCATGACTTTGT--GAAGTA--TTA-TGCTTC--GTAAA-TGG-----TTTAAAC-------A-CTTGG-TGATTAG-GTCAGT----------ATT--------ACTCCAGC---CTG-----GCTGGAGT-------------CAGATAGGCCGGCAACGGATCAATCTG-GCTGA-TTTAGCTCTGGTGCTTATCGAGGATTGGACAGGCGTA-GGCAT-GCGCAGAGAGAG-CCGGGTTGACACCGACTT------
#=GS VanFra/1-277 DE BEDY01000007.1:c723647-723353 Vanrija fragicola DNA, scaffold: scaffold_7, strain: JCM 1530, whole genome shotgun sequence
VanFra/1-277 CACCTGTAGTGGGT--TTGGGAAGGTG-TTTATT--TCCATCACGGAATTCCTTGTCTCAGCGC-TGCCC-ATTGTACCCTG--A----ATCTCCTCT-AGCACCAGTGT-TCGAGCC----CCACCAAAGTCTCACGACTTTTG----------CGA---------------TGG-----TCTTGTT---------CTCGG-TGATCAA-GCTAGT-----A-----C---------GTC--------ACGT---------GAC----AC---T---CAGATAGGCCGGTAACGGATCAGTCTG-GCTAG-TTTGGCATTGATGCTCCAAGAGGGTTGGATCGGGGTA-GGC---GCGTAGGGCGAG-CCGGATAGGCACCAACTTTTT---
#=GS VanHum/1-277 DE QKWK01000006.1:c100777-100460 Vanrija humicola strain CBS 4282 CBS4282_scaffold06, whole genome shotgun sequence
VanHum/1-277 CATCTGTAATGGAT--TTGGGAAGGTG-TTCATT--GTCATCACGGAACTCCTCGCCTCAGCGC-GCC-C-TTTGTACTTC--ATT---GTCTCCTCT-AGCGCCAGTGT-TCGAGCC----CCACCAAAGTCTCACGACTTTG-----------TGT---------------TGG------TTTC---------GTCTCGG-TGATCA-TGCTAGT-A---T-----A---------CGCC-------CTCGT-------GGCG-----G-------CAGACAGGCCGGCAACGGATCAGTCTG-GCTAG-TATGGCATTGACGCTTCAAGAGGATTGGACGGAAGTA-GGC---GCGCAGGGTGAG-CCGGGTGGACACCGACTTTTT---
#=GS CutOle/1-273 DE QKWL01000001.1:c592950-592650 Cutaneotrichosporon oleaginosum strain ATCC 20508 ATCC20508_contig01, whole genome shotgun sequence
CutOle/1-273 GCACTGTAATGGTT--TCGGGAAGATG-TTCGTT--TTCATAACGGAACTCCTCGTCTCAGTGC-GCT-C-CTTGTACAGC--CTT---GTCTCCTCT-AGCTCGAGTGT-TCGAGCC----CCACCGAGTCCTCATGGATTTG-----------TGT---------------TGG------TACC---------GCCTCGG-CAATCA-TGCCAG------------AA--------TACT-------TCCG--------GGTA-------------CAGATAGGCCGGAAACGGATCAGTCTG-GCTGG-TGTGGCATTCGTGCTCCAAGAGGTCAGGACAGCTGTA-AGC---GCGCAGGGCGAG-CCGGGCGGGCATCAACCTTTT---
#=GS CutDer/1-273 DE JAMALL010000008.1:c176229-175947 Cutaneotrichosporon dermatis strain M4303A ctg_8, whole genome shotgun sequence
CutDer/1-273 TCGCTGTAATGGCT--TTGGGAAGGGG-TTTGTT--CACATCACGGACTTCCTCGCTTCAGTGC-GCT-C-TCAATATGGCC--T----GTCTCCTCT-AGCTCCAGTGT-TCGAGCC----CCACCAAGTCCTCATGGATTTG-----------CG---------------TTGG------TCAC---------GTCTCGG-TGATCAA-GCCAGT-----------AT--------TACC-------TAAG--------GGTA-------------CAGATAGGCCGGCAACGGATCAGTCTG-GCTGG-TTTGGCATTGGCGCTTCAAGAGGTTAGGACGGTTATA-AGC---GCGCAGGGCGAG-CCGGGCAGGCTCCAACTTTTT---
#=GS DipFer/1-274 DE LFTX01002754.1:934-1219 Dipodascus fermentans strain CICC 1368 contig_6182, whole genome shotgun sequence
DipFer/1-274 TCGCTGTAATGGCT--TTGGGAAGGGG-TTTGTT-CACCATCACGGATTTCCTCGCTTCAGTGC-GCT-C-TCAATATGGCC--T----GTCTCCTCT-AGCTCCAGTGT-TCGAGCC----CCACCAAGTCCTCATGGATTTG-----------CG---------------CCGG------TCAC---------GTCTCGG-TGATCAA-GCCAGT-----------AT--------TACC-------TAAT--------GGTA-------------CAGATAGGCCGGCAACGGATCAGTCTG-GCTGG-TTTGGCATTGGAGCTTCAAGAGGTTAGGACGGTTATA-AGC---GCGCAGGGCGAG-CCGGGCAGGCTCCAACTTTTT---
#=GS DipFer/1-271 DE LFTX01003351.1:22743-23040 Dipodascus fermentans strain CICC 1368 contig_8318, whole genome shotgun sequence
DipFer/1-271 CCACTGTAACGGTT--TTGGGAAGGTG-TTTGTC--TTCATCACGGACTTCCTCGCTTCAGCGC-GCT-C-TCAATATAGCC--A----GTCTCCTCT-AGCTCCAGTGT-TCGAGCC----CCATCAAATCCTCACGGATTTG-----------CG---------------TTGG------TCAC---------GTCTCGG-TGATCAC-GCCAGT---------------------TACC-------TTCG--------GGTG-------------CAGATAGGCCGGCAACGGATCATTCTG-GCTGG-TGTGGCATTGGCGCTCCAAGAGGATAGGACGGTTGTA-AGC---GCGCAGGGCGAG-CCGGGCAAGCTCCGACTTTTT---
#=GS CutCur/1-274 DE NIUX01000020.1:c304230-303945 Cutaneotrichosporon curvatum strain ATCC 10567 Contig020, whole genome shotgun sequence
CutCur/1-274 TCAGTGTAACGCTT--TCGGGAAGGTA-TCTATT--AGCATCACGGAATTCCTCGCCTCAGTGC-GCC-C--TTAAGCAGCT--TT---GTCTCCTCT-AGCTCCAGTGT-TCGAA--TC--CCACCAAAGTCTCAAGACTTT------------TC---------------GTGG----TCACACTC---------TTCGG-TGATCAA-GCCAGT-----------AT--------TTCC-------CTCG--------GGAG-------------CAGATAGGCCGGCAACGGATCAGTCTG-GCTGG-CTTGGCATTGGAGCTTCAAGAGGTCGGGACGGCTGTA-GGC---GCGCAGGGCGAG-CCGGGTAGATACCGACTTTTT---
#=GS CryCur/1-274 DE BCJH01000007.1:c249750-249451 Cryptococcus curvatus DNA, scaffold: scaffold_6, strain: JCM 1532, whole genome shotgun sequence
CryCur/1-274 TCAGTGTAACGCTT--TCGGGAAGGTA-TCTATT--AGCATCACGGAATTCCTCGCCTCAGTGC-GCC-C--TTAAGCAGCT--TT---GTCTCCTCT-AGCTCCAGTGT-TCGAA--TC--CCACCAAAGTCTCAAGACTTT------------TC---------------GTGG----TCACACTC---------TTCGG-TGATCAA-GCCAGT-----------AT--------TTCC-------CTCG--------GGAG-------------CAGATAGGCCGGCAACGGATCAGTCTG-GCTGG-CTTGGCATTGGAGCTTCAAGAGGTCGGGACGGCTGTA-GGC---GCGCAGGGCGAG-CCGGGTAGATACCGACTTTTT---
#=GS TriGue/1-276 DE BCJX01000019.1:c2742-2450 Trichosporon guehoae DNA, scaffold: scaffold_18, strain: JCM 10690, whole genome shotgun sequence
TriGue/1-276 CACCTGTGACGGGT--TGAGGAAGGTG-TTTATT-TCCATTCACGGAAT-CATCTCTTCAGCGC-GCC-C-ATTGTACGAC--ATTA--GTCTCCTCT-AGGTCCAGTGT-TCGATCC----CCACGCGAGCTTCACGGCTCG-----------ATGCT--------------TGG------TTTC---------GTATCGG-TGATCAA-GCCAGT---------------------TGCC-------CATGT-------GGCG-------------CAGACAGGCCGGCAACGGATCAGTCTG-GCTGG-TTTGGCATTGGCCCTCCAAGAGGTCGGGACAGTCGTA-GGC---GCGCAGAGAGGG-CCGGGTAGGCACCAACTTTTTT--
#=GS PasPL2/1-275 DE JAMFRE010000099.1:c90142-89853 Pascua sp. PL2904B ctg_99, whole genome shotgun sequence
PasPL2/1-275 TACCTGTGACGGGT--TGGGGAAGGTG-TTTATT-TCCATTCACGGAAT-CACCTCTTCAGCGC-GCC-C-ATTGTACGAC--ATTA--GTCTCCTCT-AGGTCCAGTGT-TCGATCC----CCACTCGAGCTTCACAGCTCG-----------CTGCT--------------TGG------TTTC---------GTATCGG-TGATCAA-GCCAGT---------------------TGCC-------CACGT-------GGCG-------------CAGACAGGCCGGCAACGGATCAGTCTG-GCTGG-TTTGGCATTGGCCCTCCAAGAGGTCGGGACAGTCGTA-GGC---GCGCAGAGAGGG-CCGGGTAGGCACCAACTTTTT---
#=GS TriVee/1-277 DE BCKJ01000002.1:c3636532-3636236 Trichosporon veenhuisii DNA, scaffold: scaffold_1, strain: JCM 10691, whole genome shotgun sequence
TriVee/1-277 TCGCTGTAATGGCT--TTGGGAAGGTG-TTTGC--GCTTGTCACGGAACTCCTTGCTTCAGCGC-GCC-C-CTCGTACAACT--TT---GTCTCCTCA-AGCACCAGTGT-TCGAGCC----CCACCCGAGTCTCACGATTCGG----------CTGA---------------TGG------TTTC---------GTCTCGG-TGATCAA-GCTAGT-----------------GTC-CGCC-------CGCGT-------GGCG-------------CAGACAGGCCGGCAACGGATCAGTCTG-GCTAG-TTTGGCATTGGTGCTCCAAGAGGACTGGACGGTTGTA-GGC---GCGCAGGGCAAG-CCGGGTAGACACCAACTTTTT---
#=GS FusOxy/1-282 DE WESS01000049.1:c45518-45220 Fusarium oxysporum strain EtdFoc-17 scaffold68.1:size93250, whole genome shotgun sequence
FusOxy/1-282 AAGCTGTAATGGCC-CACGGGAAGATT-TCCATT-TACCAAAACGGATAAACCTTCATCAGCGC-TCC-A-TCATTACAACT--CC---GTCTCCTCT-AGCACGAGTGT-TCGAGCC-C---CACCCAGTTTTCATGAATTGG----------TTGT---------------TG-------TTTC---------G-CTCGG-TGATCAA-ACGAGT-----T----CAA-------GCGTC-------TTAC--------GGCGC----G-------CAGATAGGCCGGCAACGGATCATTCTGGCTCGGAT-TGGCATTGGCGCTTCAAGAGGATGGGACGGGTGTA-GGTA--GCGTAGTGAGGT-CCGGGTGGTTATCGACTTTTT---
#=GS NaeAur/1-307 DE JAKFAO010000007.1:981204-981525 Naematelia aurantialba strain NX-20 Contig7, whole genome shotgun sequence
NaeAur/1-307 CAGCTGTAATGGCT-TTTGGGAAGGTG-GTCTCT-TTGCATCACGGAGTTCCCCTCCTCCGAGC-TCC-C-ACATTACGATTTAATA--GTCTCCTCT-AGCACCAGTGT-TCGAGCC----CCACCCGTGTCTCACGGCGCGG---------TCTGTT--------------TGG-----CACAC---------G-CTCGG-TGACCAATGTCAGT-A---T-----A---CGCTTTGTCTCTTCCC-TCCT-GGGGGGCGACAGCG------G---CGGACAGGCCGGCAACGGATCAGTCTG-GCTGA-TTTGGCATTGGTGCTTCAAGAGGATAGGACGGTCGTA-GGA---GCGCAGGGAGGG-CCGGGTGGCCACCAACTTTTT---
#=GS SaiPod/1-298 DE RSCD01000017.1:c306302-305990 Saitozyma podzolica strain DSM 27192 scaffold_17, whole genome shotgun sequence
SaiPod/1-298 TCGCTGTCATGGCT-TTCAGGAAGATG-TCCATT-TCATCTCACGGAC-CCCCTTCCCCCGCGC-GAC-G-GGTTTATGCC--ATTC--GTCTCCTCT-AGCACCAGTGT-TCGAGCG----CCACCCCGACCTCACGGGCGGG---------TTTTAAT-------------TGG-----TCGTTT--------CACTCGG-TGATCAA-GCCAGT-----T---------CTA---CGTTTCGT--GTAAAAA--GCGAAAACG--------G---CAGGCAGGCCGGCAACGGATCAGCCTG-GCTGG-TTTGGCATTGCTGCTTCAAGAGGATTGGACGGGCATA-GGAC--GCGCAGGGAGGG-CCGGGTGGGCACCGACTTTTT---
#=GS TakKor/1-298 DE BCKT01000002.1:c694815-694510 Takashimella koratensis DNA, scaffold: scaffold_1, strain: JCM 12878, whole genome shotgun sequence
TakKor/1-298 TAGCTCTAATGGCTCATTGGGAAGATG-CCTGTTTTCCGTCTACGGATTTCCTCTCTTCAGCGC-GCC-A-TTTGTACACC--AAT---GTCTCCTCT-AGCGCGAGTGT-TCGGG--CC--CCAGCAAAGTCTCACGACTTT-----------TGTCT-------------TTGG-----TTCAGTT---------CTCGG-TGATCAT-TCCAGC-A---T-----A---CGTTTACGTAAC-----CTCG-------GTTGCGAC--G---G---CGGGCAGGCCGGCGACGGATCAGTCCCGACTGGA-ATGGCATTCGCGCTACAAGAGGATAGGACGGGCGTA-GGAT--GCGCAGAGAGAG-CCGGGCGGGCGTCGACTTTTT---
#=GS TriLai/1-277 DE BCKV01000005.1:c1767824-1767532 Trichosporon laibachii DNA, scaffold: scaffold_4, strain: JCM 2947, whole genome shotgun sequence
TriLai/1-277 CATCTGTAATGGAT--TCGGGAAGGTG-TTTGC--AGTTGTCACGGAACTCCTAGCTTCAGCGC-GCC-C-CTCCTACAATC--CT---GCCTCCCCT-AGCACTGGTGT-TCGAG--CC--CCACCAGAGTCTCACGACTCTG-----------TGA---------------TGG-----TTTCGTT---------CTCGG-TGATCAA-GCCAGT----------TCT--------CCCC-------CACGT--------GGGG------------CCGTCAGGCCGGAAACGGATCAGACGG-GCTGG-TTTGGCATCAGTGCTCCAAGGGGATTGGGCGGTTGTA-GGC---GCGCAGAGCTAG-CCGGGCGAGCACCAACTTTTT---
#=GC Domains -------2------|------------5-a-----|--------5b|-9--|--5c-------------------|-10------|-------5d---|-----5e----|-6a-------6b----------6c---------------------------------------6b----------------------6a--|-------8a-----|--------------------------------8b-------------------------|------------8c-------------|---8a-----|----5e---|------5d-------|--11-|-------5c--------|-5b--|--5a--------5a--|---
#=GC SS --(((-----)))---(((((--((((-((((((---------(((-----(((((((--((((-(((-------((((((--------(((((((((-(((-(((((((-(((((-(---((((-(((((((----)))))))-----------------------------))))------------------)-)))))-----(((-((((((--------------------((((((-----------------))))))------------((((--(((((----)))-))--))))-)))))-)))))))))))-)))---))))))---)))-))))))-)))---))))-)))))))-)))))))))))))--)))))----
//
Image source Saccharomyces’ SRP RNA:
# STOCKHOLM 1.0
#=GF SS 2D_Scer From Suppl. Figure S1 https://doi.org/10.1016/j.jmb.2007.02.056
#=GS SacCer-CP092957/1-523 DE CP092957.1:c144246-143724 Saccharomyces cerevisiae S288C chromosome V
SacCer-CP092957/1-523 -AGGCTGTAATGGCTTTCTGGTGGGATGGGATACGT-T--GAGAAT--TCTGGCCGAGGAACAAATCCTTCCTCGCGGCTAGA------CACGGAT-TGCAC---GCCC---------------TTT--------------GGGCAAGG------GATAG--TTCT-CTATTCC--------------------------------------------GCACCGTG------CCCTGTTGTGGCAACCGTCTTTCCTCCGTCGTA-AATTTG-----TCC-TGGGCAGA----GCTG---TCTGCCCGG--AGGCGGGAGAGTCC----GTTCT--GAAGTGTCCCGGCTA---TAATAAATCGATCTTTGCGGG-CA-----GCC---CGTTGGCAG------GAGGCGTGAGGAATCCGTCTCT-CTGTCTGGTGCGGC-AAGGT------AGTTC-TGG---------GTCC------TTA-GGGGCT-------C---CACCTTCACCGCTGTTAGGGGAGTTTTATCCAGCGTCAGCAAAGGTG-ACCCGTGATGGAGGCGG-CCGGG--ATAGCACATATCAGTCGGATAAT-TGTGCAAGTTG-A-TCGCTTCGGCGGTTTA-ATTT--GGCGGTGCCATCAGG----ATTTACTCGCACATTGTGGCCGTTCCCTCGGGGATGGA--GTGTGTCC--TGAACCAT------ATTTTTT------------
#=GS SacPar-BN000251/1-520 DE Saccharomyces paradoxus scRNA
SacPar-BN000251/1-520 -AGGCTGTAGTGGCTTTCTGGTGGGATGGGATACAT-T--GGGAAT--TCTGGCCGAGGAACAAATCCTTCCTCGCGGCTAGA------CACGGAT-TGCAC---GCCC---------------TTT--------------GGGCAAGG------GGTGG--TTTT-CTACTCC--------------------------------------------GCACCGTG------CCCTGTTGTGGCAACCGCCTTTTCCCTGTCGTA-AATTTG-----TCC-TGGGCAGA----GCTG---TCTGCCTGG--AGGCAGGGGAGTCC----GGTCT--GAAGTGTCCCAGCTA---AAATAAATCGATCTTTGCGGG-CA-----GCC---CGTTGGCAG------GAGGCGTGAGGAATCCGTCTCT-CTGTCTGGTGCGGC-AAGGT------AGTTC-TGGG---------TCC-----CTTA-GGGGCT-------C---CACCTTCACCGCTGTTAGGGGAGTTTTATCCAGCGTCAGCAAAGGTG-ACCCGTGATGGAGGCGG-CCGGG--ATAGCACATATCAGTCGGATAAT-CGTGCAAGTTG-A-TCGTTTCGGCGGTTTA-ATTT--GGCGGTGCCATCAGG----ATTTACTCGCACATTGT-GCCATTCCCTTGGGAATGA---GTGTATTC--TGAACCATT------TTTT--------------
#=GS SacBay-AJ550801/1-526 DE Saccharomyces bayanus scR1 101..624
SacBay-AJ550801/1-526 -AGGCTGTAATGGCTTTCTGGTGGGATGGGATACGT-T--GGGAAT--TTTGGCCGAGGAACAAATCCTTCCTCGCGGCCAGA------CACGGAC-TGCAC---GCCC---------------TTT--------------GGGCAAGG------GATGG--TTCT-CCATCTC--------------------------------------------GCACCGTG------CCCTGTTGTGGCAACCGTCTTTTCTCCGTCGCT-AATTTG-----TCC-TGGGCAGA----AATG---TCTGCTCGG--AGGCGGGGGAGTCC----GGTCT--GAAGTGTCCCGGCTA---TAATAAATCGATCTTTGCGGG-CA-----GCC---CGTTGGCAG------GAGGCGCGAGGAATCCGTCTCT-CTGTCTGGTGCGGC-AAGGT------AGTCC-TGGG--------TCCC-----TTT--GGGGCT-------C---CACCTTCACCGCTGTTAGGGGAGTTTTATCCAGCGGCAGCAAAGGTG-ACCCGTGATGGAGGCGG-CCGGG--ATAGCACATATCAGTCGGATAAT-CGTGCAAGTTG-A-TCGTTTCGGCGGTCTA-ATTT--GGCGGTGCCATCAGG----ATTTACTCGCACATTGTGTTCGTTCCCTCGGGGACGA---GTGTGTATCCTGAACCAC------ATTTTTTT-----------
#=GS SacKud-AJ550803/1-526 DE Saccharomyces kudriavzevii scR1 101..623
SacKud-AJ550803/1-526 -AGGCTGTAATGGCTTTTCGGTGGGATGGGATACAT-T--GGGAAT--TCTGGCCGAGGAACAAATCCTTCCTCGCGGCTAGA------CACGGAT-TGCAC---GCC---------------TTTC---------------GGCAAGG------GATAG--TTTT-CTATTCC--------------------------------------------GCACCGTG------CCCTGTTGTGGCAATCGTCTTTCTCCCGTCGTC-AATTTG-----TCCCCGGGCAGA----ACTG---TCTGCTCGG--AGGCGGGGGAGTCC----TATCT--GTAGTGTCCCGGCTT---TAATAAATCGATCTTTGCGGG-CA-----GCC---CGTTGGCAG------GAGGCGTGAGGAATCCGTCTCT-CTGTCTGGTGCGGC-AAGGT------AGTTGT-GGG---------TCC-----CTT-TGGGGCT-------C---CACCTTCACCGCTGTTAGGGGAGTTTTATCCAGCGTCAGCAAAGGTG-ACCCGTGATGGAGGCGG-CCGGG--ATAGCACATATCAGTCGGATAAT-CGTGCAAGTTG-A-TCGTTTCGGCGATTCA-AGTT--GGCGGTGCCATCAGG----ATTTACTCGCACATTGT-GCCATTCCCTCGGGGATGGT-TGTGTGTTC--TGAACCAC------ATTTTTTTT----------
#=GS SacMik-AJ550802/1-525 DE AJ550802 1-525 Saccharomyces mikatae
SacMik-AJ550802/1-525 -AGGCTGTAATGGCTTTCTGGTGGAATGGGATACGT-T--GAGAAT--TTTGGCCGAGGAACAAATCCTTCCTCACGGCCGGA------CACGGAT-TGCAC---GCCC---------------TTT--------------GGGCAAGG------AGTAGA-ATT-TCTATTTC--------------------------------------------GCGCCGTG------CCCTGTTGTGGCAACCGTCTTTCCCCCGTCGTT-GGTTTG-----TCC-TGGGCAGA----GCTG---TCTGCCCGG--AGGCGGGGGAGTCC----TATCT--AAAGTGTCCCGGCTT---AAATAAATCGATCTTTGCGGG-CA-----GCC---CATTGGCAG------GAGGCGTGAGGAATCCGTCTCT-CTGTCTGGTGCGGC-AATGT------AGTTC-TGGG---------TCC-----CTTA-GGGGCT-------C---CACCTTCACCGCTGTTAGGGGAGTTTTATCCAGCATCAGCAAAGGTG-ACCCGTGATGGAGGCGG-CCGGG--ATAGCACATATTAGTGGGATAAT-CGTGCAAATTT-A-TCGCTTCGGTGGTTTA-AATT--GGCGGTACCATCAGG----ATTTACTCTCACATTGTGGTCATACCTTCTGGTGTGAG-TGTGTATTC--CAAACCATT------TTTTT-------------
#=GS SacMik-BN000249/1-525 DE BN000249 1..525 Saccharomyces mikatae
SacMik-BN000249/1-525 -AGGCTGTAATGGCTTTCTGGTGGAATGGGATACGT-T--GAGAAT--TTTGGCCGAGGAACAAATCCTTCCTCACGGCCGGA------CACGGAT-TGCAC---GCCC---------------TTT--------------GGGCAAGG------GGTAGA-ATT-TCTATTTC--------------------------------------------GCGCCGTG------CCCTGTTGTGGCAACCGTCTTTCCCCCGTCGTT-GGTTTG-----TCC-TGGGCAGA----GCTG---TCTGCCCGG--AGGCGGGGGAGTCC----TATCT--AAAGTGTCCCGGCTT---AAATAAATCGATCTTTGCGGG-CA-----GCC---CATTGGCAG------GAGGCGTGAGGAATCCGTCTCT-CTGTCTGGTGCGGC-AATGT------AGTTC-TGGG---------TCC-----CTTA-GGGGCT-------C---CACCTTCACCGCTGTTAGGGGAGTTTTATCCAGCGTCAGCAAAGGTG-ACCCGTGATGGAGGCGG-CCGGG--ATAGCACATATTAGTAGGATAAT-CGTGCAAATTT-A-TCGCTTCGGTGGTTTA-AATT--GGCGGTACCATCAGG----ATTTACTCTCACATTGTGGTCATACCTTCTGGTGTGAG-TGTGTATTC--TAAACCATT------TTTTT-------------
#=GS SacCas-AJ550804/1-549 DE AJ550804 101-633 Saccharomyces castellii
SacCas-AJ550804/1-549 -AGGCTGTAATGGCTTT-TAGTGGAACCAAGTATTTT---GGGATA--TTTGGCCTGGGAA-CGAAACTTCCTTACGGTCAGA------CATGGAT-TGCAC---GTTTCGTC-----------TTT----------GGCGGAACAAGGGGTTTTGCCAA--GTT-TTGGTA-GGGTTCT--------------------------------------GCGCCATG------CCCTGTTGCTTCAACCGTCTTCCTGAACCCGCCTGTTAGGGCTTGTC-TCGGCTGGTGTAGTTTTCTGTGCCAGTTGGAGGGTTTAGGGCTCT----GAGTT--AACGTGTCCCGGTTTA-ACAATCAATCGATCTTCGCGGG---TT---GCT---CGTTGGCAG------ACAGCGCGAGAAATCCGTTGTTTCTGTCCGATGCGGC-AATGTAATCT-------GGTC--------ATGT----GACC------------------TCGCCTTGACCGCCATTAGGGGAGATTCATCCAGCGATGGCAAAGGCG-ACCCGTGAGAGAGGCGA-CTGGG--ATGGCAC-TGTTCTTAGGGTTAACAAGT-------ACAAGTTCG-----GTTTA-TTAT--GGACGGTGGAGTCTGG---TATTTCCTACCACATGTTGG-----------GTGT---AAACTACTCTTAGGAACCACTAACTACTTTATTTCTTTTTT----
#=GS CanGla-HG323640/1-504 DE >HG323640.1 TPA: Candida glabrata SRP RNA for signal recognition particle RNA
CanGla-HG323640/1-504 -AGGCTGTAATGGCTATTTAGTGGAATTGTTCTCCT-T--GGGAAT--TTTGGTCTGGGT-TCGA----ACCCTACGGCCAGA------CGCGGA--GGCAC---GCCTTTAATTATTACACCGTTTCGGTGGGGGCAAAGGAGCAA--------GCTGGG-CTCG--CCCAGCC------------------------------------------TGT-TCCTG------CACCGTGACTT------------CCCGTGAACTGCAGTCGTC--TCCCC------------CCTGT--GGGGA-----AGGC-----GAGCCACCCAGG-----GACGTGTCTCGGTTCATCCAACCAATCGATCCTTGCGGG-C------GTTCG-TGGCCCCTG---CTGGCGCTGTGAGTAATCCAGTG-TCCTCTAGGGC-CGGC-AACGTA-GCTACGGTCTACCCTCC------CTA------GGAGGGGGGC----------CCACCTTCACCGCCGTTAGGGGAGTTTCATCCAGCGATGGCAAAGGTG-ACCCGTGATGGAGGCGG-TCGGG--ATGGCACTGGTCCCGTGGTT---------------ACCTACCTGC----------------AGTCGCGGTG------GC-ATTATCCT-GCCCTTCAATGGGC----TTT---------GGGATT--ATCAAGACCAC-----AATTTTTTTTT---------
#=GS SacKlu-AJ550805/1-453 DE (Saccharomyces) Lachancea kluyveri SRP RNA 101..539
SacKlu-AJ550805/1-453 AGAGCTGTAATGGCTTT-TGGTGGAACCAGGTGTCCC---GAGAGT--TCTGGTCTGGGT-TCGA----ACCCTACGGCCAGA------CGTGGT--TGC----------------------AATTATA-------------------------CAACTC-TTTGT--GGGTTG--------------------------------------------GCGCCGCG-------CCCA--AGCTCAACTGC-A--------CTT--------------CCTC------------GTAA---GAGG-------AG---------CCG----AGTTTTAGACGTGTCCCGGATT-------ATATCGATCTTTGCGGG-GA-----GT-------CCTGGGCG-----AGACATGAGAAGTCTGTCT-T-CGCTTAGGTACGGC-AATGGT---ACCACACGACG------CACCTTGCGT---CGTTGTGTGGTTTTCG-CA--TGCTCCTTCCGCTG-AAGGGGAGTTTCATCCAGCA-CAGCAGGAGT-CACCCGTGA-GGAGGCGG-CCGGG--ATGGCACTCGTCCTCGGTAT---------------CATTAAGCAC-----------------GGCGGTGG-GCTGGG---ATTTCCTCG--GGGCATTT----AGCCTCTGTT-------GGACACCTTAGGAACTACCAAAGTATTCTTTTTTTTT------
#=GS VanPol-AAZN01000244/1-464 DE >AAZN01000244.1:271176-271639 Vanderwaltozyma polyspora DSM 70294 KpolContig1002a, whole genome shotgun sequence
VanPol-AAZN01000244/1-464 -AGGCTGTAATGGCTTCATGGTGGAACTGT--TAACCTT-GAGAAT--TCTGACATGGGA-TCAAT---TCCCAACGGTCAGA------CACA-TGTCACTA---ACCG-TC------------TTT----------GAGCGGTGAAGT------TGCGT---CC---GCGTAGCT------------------------------------------GTGCTGTG-------TTCG--ATGGTATTTCAGTCG-------------------------TT------------TTAGC--AA----------------------------------GATGTGTCCCGGTT--AAAATTTAATCGATCCTTGCGGG-A------GTTGT-CCGATATCGGT-----CGGCATGAGAAGTCTGTCGTATCGTA-CGGTACAGC-AATGTAATCAATT-GTGTTCTTCC----CCTCAAAA---GGAAG-GTGC--------T---GCTCTTACCGTTATTAGGGGAGTTTCATCCAGCAATGACAAGAGC-TACCCGTAATGGAGGCGG-TCGGG--ATTGCAC-TGTCTGCTTC-----GTGGTCAG------TTCT----CTGATCCAC--------ATCGACTG-CCGT-GAA-ATTATCTC---TTCT---CGTCTAACGGGA-TATGG--GGTTATTCTTCAGGAACCACCGC---ATTT---------------
#=GS KluWal-AADM01000319/1-418 DE AADM01000319.1:21425-21842 Lachancea waltii NCYC 2644 cont318, whole genome shotgun sequence
KluWal-AADM01000319/1-418 ATCACTGTAACGGTCGACA-GTGGAACTGTATTTTC-C--GAGATT--TCTGACCTGGGT-TCAA----ACCCTGCGGTCGGA------CGTGTTAC-GC------------------------TTTA-------------------------AGCCTCT--TTCG--GGAGGTT-------------------------------------------GCTACACG------CTCAGTCT-AACAAT----------------------------------------------------------------------------CG----AGTC---GACGTGCCCTGGT------AACAAATCGATCCTTGCGGG-CA-----GTC---CAGGGC---------GAGATGTGAGAAGTCCGTCTT--GTACCTGGTACGGC-AATGGT-----AGGCGAGGGTG-------CTTAA-----CGCTA-ACCCG-TCT-CTT-GTGCTCCTATCGCTG-AAGGGGAGTTTCATCCAGCA-CAGCAAGAGCA-ACCCGTGATGGAGGCGA-CTGGG--ATTACACTTGTCCTCG----------TATAAGGGA---TTTA---TCCCTT-------C---GTTGTGGGC-----GTG-AAT--CTC-ATCCTATGGA-------ATGT----------GAA-GATACTAGAACCCTGTC---ATATTATTTTTTA------
#=GS ZygRou-AJ564197/1-533 DE AJ564197 1-533 Zygosaccharomyces rouxii
ZygRou-AJ564197/1-533 --AGCTGTAATGGCTCAATGGTGGAATTGTACACCT-T--GGGATT--TCTGACTTTGG-TTCAAT----CCTCGCAGTCGGA------CGTAGGTTAGCA----TGTCACGT-----------TTCG----------GCGTGGCAAGGGGTA--CTCAC-CTTCCC-GTGGGG-CTCC---------------------------------------GCACTGCG------C--GTCGGCGACAACCGTCATCCTTCTCTCGCT-TCCAGT---TGCC-GAGGCCGTCA---ATCT---TGGCGGTCTCAGAGAGAGGGACTCG----GGGTT--AACGTGTCCCGGTTA-------TTATCGATCTTCGCGGG-G---TTTGTT------TGCGCGTTCGGG--CACGAGAGAAATCCGTGTT--CCTGGGCGTGTGGC-AAAGTA-------GTTACGGG---------TTC----CTTGTGGGC----------C--TCGCCTTGACCGCTG-AAGGGGAGTTTCATCCAGCA-CAGCGAGGGCG-ACCCGTGATGGAGGCGG-CCGGG--ATAGCACTAGTTCTCCGAGTACAAGTGCAGACTGGGTTGGCCCTGTGCCAATCTTAACG---GGCGGTGTCGCTGAT---AATTACCCACCAGTAAAGGT-ACGTTGCTTGCAGCGTG--GGGTGTAC--AAAACTATT------TT----------------
#=GS ZygBai-HG316458/1-525 DE HG316458.1:c717351-716827 Zygosaccharomyces bailii CLIB 213 genomic, scaffold05
ZygBai-HG316458/1-525 --AGCTGTAATGGCTTT--GGTAGAACTGTACGCTTC---GAGATT--TTCGGCTTGGG-TTCAAT----CCTTACGGTCGGA------CGTAGGTTAGCG----TGTCATGC-----------TTCG----------GTGTGACAAGGGGTT--CCTGT--GAAA--GCAGGAATTCT---------------------------------------GCACTGCG------C--GTCGGTGCCAACCACTAT-CCTCTCCC-TGTTTCCA--TTTGC-GGAGGCGACT--AGCCTATTA-GGTTACCTCTGGGGGAGG-ACCTG----GAGTT--AACGTGTCCCGGTAC-------TTATCGATCTTTGCGGG-----TTTGC-------AGAGC--CTAGG--CACGAGAGAAGTCCGTGA---TCTAG-CCTGTGGC-AACGTA--------GTATGGG---------CTC----TTTGTAGC-----------C--TCGTCCTCACAGCTG-TAGGGGAGATTCATCCAGCA-CAGCAGGGGCG-ACCCGTGACGGAGGCGG-TCGGG--ATAGCACTAGTTCTCCGGGTATTAGTGCAGTGTGGGCTCGCTTGTCGGGTAACATTT-----GGTGGTGGCACTGAT---AATTGCTCCTTTGTTAAG-CCTCTGGTTCTCCAGTGG--GGAGCGTAT--TGAACTACT------TTTTT-------------
#=GS EreGos-HG323639/1-381 DE HG323639 Ashbya gossypii ATCC 10895 SRP RNA for signal recognition
EreGos-HG323639/1-381 --AGCTGTGACGGCCTTTGGCCGGAAGGGACATTA-G-----CAA--TCCCGGCTTGGTGAGGCGCTCCCGCGTTCGAACCTGCGGCCGGGCATGGTTGC-----------------------GGAAAA----------------GCGAGGTT-----GC---CTA--GCTCCTC-----------------------------------------GTGCGCCATGA-----------TACCACACTGGTCT-------------------------------------------TT--------------------------CT----GA-----CGCGTGTCCTGCT-------ACTTATCGATTTCTGCGGG--TTTGCCGT----------CGATGCT---GA-CATGGGAAACCTGTC-----GGTGGGGCGTG-C---AGTA-----ATGGCA-----------GGGCGAA--TGCCCGT----------------CCGCTCATACGCTG-GAGGGGAGTTTCATCCAGCACAGCGCGAGCGG--CCCGTGG-GGAGGCGAGCAGG---ATGGCAC--GTGTTAG---------------------GCGCTTGCGC---------------GGACCTATCGTGGTA----TTG--------GAGCA------------------------TTATGTT--CCGACCGACT-----TTTT--------------
#=GS KluLac-HG323642/1-496 DE >HG323642.1:1-496 TPA: Kluyveromyces lactis SRP RNA for signal recognition particle RNA
KluLac-HG323642/1-496 ---------------ACCCTGTCGAAGGAGTGTGATG---CTTGGATATGTGGCTCCGGTTCGAAACCGCACAGTCACA----------CCGGGT----------CTCTTTCAAATGGAAGAGGTTTCACAAT-GGTATC-CCAAACAGGCGTGAAATGTCTC---------------GTAGTGGTTATATGCCATTACCCGCTTTTCGT--GTCTGGCCGCCCGG------------TCTACGCG----------------TTCATTTATTTGTGTAACTGCTTTGGACAGCTGGATCTCGGTTCGG---------------TTGTCACGTGGTCGACGTGTCCCACCGTT-------ACAAAATTGTTGCGGGTTATCCTTTG---------TGGTGGCTCGGTCGCATGAGAAGTCTGCGGTTCGAAGCCCAACTGGTAAC------GATTTAGCATTTCAA-----CTAATAC--TTGAAGCTAGTT----------TAGTGTTTAACGTTG-TAGGGGAGTCTCATCCAGCACAACGAACACTG--CCCGTGAC-GA---GGTGGTGTGGATTGCAC--GTCCCGTGGTTAAGCAG------------------------------------TGCGTGTGG---------GTTCAGAG----GCAGA-------------------------CATACA--CTGTCCTTC-----ATTTTT-------------
#=GS KluLac-CP042456/1-500 DE CP042456.1:1273402-1273901 Kluyveromyces lactis strain CBS 2105 chromosome 2
KluLac-CP042456/1-500 ---------------ACCCTGTCGAAGGAGTGTGATG---CTTGGACATGTGGCTCCGGTTCGAAACCGCACAGTCACA----------CCGGGT----------CTCTTTCAAACGGAAGAGGTTTCACAAT-GGTATC-CCAAACAGGCATGAAATGTCCC---------------GTAGTGGTTATATGCCATTACCCGCTTTTCGT--GTCTGGCCGCCCGG------------TCTACGCG----------------TTCATTTGTTTGTGTAACTGCTTTGGACAGTTGGATCTCGGTCCGG---------------TTGTCACGTGGTCGACGTGCCCCACCGTT-------ACAAAATTGTTGCGGGTTACCCTTTG---------TGGTGGCTCGGTCGCATGAGAAGTCTGCGGTTCGAAGCCCAACTGGTAAC------GATTTAGCATTTCAA-----CTAATAC--TTGAAGCTAGTT----------TAGTGTTTAACGTTG-TAGGGGAGTCTCATCCAGCACAACAAACACTG--CCCGTGAC-GA---GGTGGTGTGGATTGCAC--GTCCTATGGTTAAGCAG------------------------------------TGCGTGTGG---------GTTCAGAG----GCAGA-------------------------CATACA--CTGTCCTTC-----ATTTTTTTTT---------
#=GS KluMar-AP014601/1-519 DE AP014601.1:c280853-280319 Kluyveromyces marxianus DNA, chromosome 3, complete genome, strain: NBRC 1777
KluMar-AP014601/1-519 -----AATTGGCCTTTCTGGAAGTCAGAGATGTGATAC---TTGGATAAGTGGCTCCGGTTCGAAACCGCACTGTCACT----------CCGGGT-----------CCCTTGTATTAGGGG---TCTCCAC---GGTACTATTAAACAGCATCGGTAGTCTCTTGTGAG-------ACCCGTCCTACTCGTAGGCGGGTAACAAGCTGCTGGTGTTGGCCGCCCGGCCCAGACGTCCCACTAGA----------ACTTCTT----CC-----------GATCTGAGAGATTTTATCGTCTCACAGGTC--------------------ACGTGGTCGACTTGTCCCACCGTT-------TCAAAATTGTTGCGGGTCTTTCCTGTA--------TGGTGGCGCGACTGCGCGAGAAATCCGCGGTTCGA-GTCCAACTGGTAACGATTTAGC---AAA--TTTTCCCAT--CTCGC---ATGGGAAAGCCCGCTAA--T--TAGTGTTTACTGTTG-AAGGGGAGTCTCATCCAGCACAACGAACACTG--CCCGTGAC-GA---GGTGGTGTGGATTGCAA--GTCCCAT-GT-------------AT------AAGGAGT----------------ACGTTTGG--------ATCCAGAT--------------------------------GTATCAGCTAGCCTGACCAATT----TTTTT-------------
#=GS KluMar-CP015056/1-519 DE CP015056.1:c304887--304369 Kluyveromyces marxianus strain FIM1 chromosome 3, partial sequence
KluMar-CP015056/1-519 -----AATTGGCCTTTCTGGAAGTCAGAGATGTGATA---CTTGGATAAGTGGCTCCGGTTCGAAACCGCACAGTCACT----------CCGGGT-----------CCCTTGTATTAGGGG---TCTCCAC---GGTACTATTAAACAGCATCGGTAGTCTCTTGTGAG-------ACCCGTCCTACTCGTAGGCGGGTAACAAGCTGCCGGTGTTGGCCGCCCGGCCCAGACGTCCCACTAGA----------ACTTCTT----CC-----------GATCTGAGAGATTTTATCGTCTCGCAGGTC--------------------ACGTGGTCGACTTGTCCCACCGTT-------TCAAAATTGTTGCGGGTCTTTCCTGTA--------TGGTGGCGCGACTGCGCGAGAAATCCGCGGTTCGA-GTCCAACTGGTAACGATTTAGC---AAA--TTTTCCCAT--CTCGC---ATGGGAAAGCCCGCTAA--T--TAGTGTTTACTGTTG-AAGGGGAGTCTCATCCAGCACAACGAACACTG--CCCGTGAC-GA---GGTGGTGTGGATTGCAA--GTCCCAT-GT-------------AT------AAGGAGT----------------ACGTTTGG--------ATCCAGATG--------------------------------TATCAGCTAGCCTGACCAATT----TTTTT-------------
#=GS KluPCH-JAGEVD010000013/1-518 DE JAGEVD010000013.1:c72437-71920 Kluyveromyces sp. PCH397 tig00000021, whole genome shotgun sequence
KluPCH-JAGEVD010000013/1-518 -----AATTGGCCTTTCTGGAAGTCAGAGATGTGATA---CTTGGATAAGTGGCTCCGGTTCGAAACCGCACAGTCACT----------CCGGGT-----------CCCTTGTATTAGGG----TCTCCAC---GGTACTATTAAACAGCACCGGTAGTCTCTTGTGAG-------ATCCGTCCTACTCGTAGGCGGGTAACAAGCTGCCGGTGTTGGCCGCCCGGCCCAGACGTCCCACTAGA----------ACTTCTT----CC-----------GATCTGAGAGATTTTATCGTCTCCCAGGTC--------------------ACGTGGTCGACTTGTCCCACCGTT-------TCAAAATTGTTGCGGGTCTTTCCTGTA--------TGGTGGCGCGACTGCGCGAGAAATCCGCGGTTCGA-GTCCAACTGGTAACGATTTAAC---AAA--TTTTCCCAT--CTCGC---ATGGGAAAGCCCGCTAA--T--TAGTGTTTACTGTTG-AAGGGGAGTCTCATCCAGCACAACGAACACTG--CCCGTGAC-GA---GGTGGTGTGGATTGCAA--GTCCCAT-GT-------------AT------AAGGAGT----------------ACGTTTGG--------ATCCAGATG--------------------------------TATCAGCTAGCCTGACCAATT----TTTTT-------------
#=GS KluDob-PPJP02000011/1-523 DE PPJP02000011.1:c238726-238204 Kluyveromyces dobzhanskii strain NRRL Y-1974 NODE_11_length_285983_cov_19.1963_ID_21, whole genome shotgun sequence
KluDob-PPJP02000011/1-523 ----------ACAGCATCCTGGCGAAGGAGATAGCAT---CTAGGACATGTGCCTCCGGTTCGAAACCGCACAGGCACA----------CCGGGT-----------CCCTTTCAATGAGAGAGGTCTACGGAT-GGTACC-CCAAACAGGGGCGGCGCTGTCACGTGTC-------CATTGGTCTAACCGCCAATTCCGCGTGCTGCACCGTTCTTGGCCGCCCGGTCTACGCGTTCATAGCTTCGTGCTTCCAACTGTTTT--G-------------GACAGCGCCA----TCC---TGGCGCTGTC--------------------ACGTGGTCGACGTGTCCCATCGTT-------TGAAAATTGTTGCGGGTTATCTTTGT----------GGTGGCTCGGTCACGTGAGAAGTCCGTGATTTGAAGTCCAACTGGTAACGATTTAGCTTCAACATCGGTCCAAT---TTC----ATTGGATTAGAAGCTAG--TT-CAGTGTTCATCGTTGT-AGGGGAGTTTCATCCAGCACAACAAACACTG--CCCGTGAC-GA---GGTGGTGCGGATTGCAC--GTCCCATGGT--------------------AAAGCAGT----------------ACGTGTGG--------GTTCACAGG-------------------------------TTGCTTACAGCCCTTC-------ATATTTTTT------------
#=GS KluSta-WACN01000003/1-528 DE WACN01000003.1:746854-747381 Kluyveromyces starmeri strain UFMG-CM-Y3682 NODE_3_length_972539_cov_72.8531, whole genome shotgun sequence
KluSta-WACN01000003/1-528 ------------ACTCCCTCTAGGGGTGCGGTCTGTC---TGCAGACATGTGACACCGGTTCGAAACCGCACGGTCACT----------CCGGGT-CTTTCTTCTGAAAGAGATTCCGCCTTCGGGCGGTTTTCGGTACTCCCATAAACAGCGAGGACGGGGCAGTTCTTGTCACGTGATTAGGTATCCTAGCCACGTGACACTGTCTCCCTCTCTGGCCGCTCGGTCAATGC--AAGCTAACG------GGAGCCTTCATC----------------GGCACAGGACGAACCTGTTCTTTCTGTGTC--------------------ACGTGGTGGACTTGTCCCACCGTT-------TGAAAATTGTTGCGGG---TAACATT---------TGGTGGTTTGGCCGCGCGAGAAGTCCGCGGTTCGAAGCCCATCTGGTAACGACC--TTGGTATT-TA-GTC-------TAAT-------GAC--GATACCAA--AC-CAGTGTTTAACGTTG-AAGGGGAGTCTCATCCAGCACAACGAACACTG--CTCGTGAC-GA---GGTGGTGAGGATTGCAA--GTCCCATGGT--------------------GAGAAGGTCC----------------GCATTG--------GTCTTAGCA--------------------------------GATGGTTAGCCGTT-----CGATTATTTTTTGTTT--------
#=GS KluWic-AEAV01000057/1-440 DE AEAV01000057.1:40798-41237 Kluyveromyces wickerhamii UCD 54-210 contig00060, whole genome shotgun sequence
KluWic-AEAV01000057/1-440 -------ACCTGTCGTGGTACAAGGGATGTGATCAGT---ATAAGATATGTGGCTCCGGTTCGAAACCGCACAGCTGCA----------CTGGGTCC-------------------------------------GGTA-TTCCAAAC-----------------------------AGCAACTTTCTTTCGGGAATTCT-------------TGTTGGCTGCCCAGCTTCTGCG-GAATCAAA-----------ACCACTT----T------------GGCTAC------AATAC-------GTGGTC--------T------------CGTGGTTGACGTGTCCCGTTGTT-------TCAAAATCGTTGCGGG--TTTATTAA-------------TGAGGCTCCACATGAGAAATCTGTGCAAAGC-GTCTATTTGGTAAT----TGTTCCGC-ATTCAGGT-------TCTAT---GCCTGAAT-CGG--TCA-----CGTGTTTACCGTTG-GAGGGGAGTTTCATCCATCACAACAAACACG-G-CCCGTGAC-GA---GGTAGCGTGGATTGCAT--GTCCCAT-G------------TCCT------AAGTGGTA----------------CGCGGAA--------AATCTTGT---------------------------------AATTCAGGCTGGACACAAACCCTTGGCTTCTTTTTTC------
#=GS KluNon-QYLQ01000002/1-421 DE QYLQ01000002.1:867865-868285 Kluyveromyces nonfermentans strain NRRL Y-27343 flattened_line_2, whole genome shotgun sequence
KluNon-QYLQ01000002/1-421 ---ACTGTTGTGGCATGGTAAAGCGGAAGCGAGCACT----TTGAAGATGTGTCCCCGGTTCAAAACCGTACGGTCA------------CAGTGG------------------------------------CATGGT-AATACAAACAA------------------------------TAACACACATTTGTGTGTTA---------------TTGGCCACCCACTG-CTCAAGTA-----------------ACCATC-----TC-----------AGTTTG----TTATTGTT------CAAGCT--------G------------CGCGGTTGACGTGTCCCATTGTT-------TCAAAATCGTTGCGGG---TGTCTAA------------GTCTGGTCACGTGTAAGTAATTCGCGT-TGATCAGCTGTTTGGCAAT----TGTA--GT-A----GG--------TTTT-----CT--------AC-GCA-----CGCGTGAACAGTTG-GAGGGGAGTTTCATCCAGCACAACGAGCGCG-G-CCCGTGAT-GA---GGTGGTGAGGATTGCAC--GTCCCGC-G------------AAAT-------GATGGT------------------ACTGGG--------TTCTTCAG----------------------------------CACGGTGTTGTTACATACCGC--ATTTCTCTTTTTCCTTTTT
#=GS KluAes-PPJO01000006/1-416 DE PPJO01000006.1:54130-54545 Kluyveromyces aestuarii strain NRRL YB-4510 NODE_6_length_597287_cov_27.2088_ID_11, whole genome shotgun sequence
KluAes-PPJO01000006/1-416 ----ACATCTGTACTGGTTTGACGGACTGGAACGGCT----TAGGATACGCGACCCCGGTTCGAAACCGCACGGTCGTG----------ATGGGT--------------------------------------CGGT-AACTCAAACAA-----------------------------AAACTCACGTCTCGTGTTT--CT-------------TTGGCCGCCCAT-GCTTCTGTTA-----------------GCCACCT-----------------TGACCCAGGC--ATTTA---GCTTTGGTCA---------------------CGTGGTAGACGTGTCCCATTGTT-------TCAAAATCGTTGCGGG---TGTCTA------------TGGCTGGTCTCGTGTGAGTAGTCCGCG-TTGGGCAGTCGTTTGGCAATT--------GC--A-ATAGG--------TTATC----CCTAT-----GT-----TA--CACGTGAACAGTTG-GAGGGGAGTATAATCCAGCACAACGAGCGTG-G-CCCGTGAT-GA---GGTGGTGCGGACTGCAC--GTCCTAC-G------------ATAT------AGGTGGT----------------ACAGTTGT--------TGTTCTG-----------------------------------TCATGTTGGTCTAAACCGT----CTTTTTTT-----------
#=GC Domains --2---------2-----|--------5a--------|----5b---|--------9a-------------------9a----------|----9b-----|----9c--------------------------------9c---|------9d-------------9d---------------------------------------------------9b----|------------5c------------|-------10a-----------------10b--------------------10b---|---10a---|------5d----------|---5e-------|--------------|--5f------|----------------------6------------------------------6----------|-----|---------7------------------------------7---------|-------8a--------8b-------------8b------------8a-|-----5f------|-----5e------------|------5d--|--11a---|---11b-----------11b---|--11a-|--------5c------|--5b------|---12a---|-------12b--------|----5a-----------5a--|-----------------------
#=GC SS_cons -(((((-----)))))----((((--((((((((((----((((((--(((((((((((((-------))))))--)))))))------(((((---(((-----((((--------------------------------))))--((------(((((-------)))))))--------------------------------------------))))))))-------((((--(((((-((((((----(((((((---------------(((--(((((((-----------)))))))))--))))))-))-((((----(--((--((-(((((((((((---------------((((((((((--------(((---(((((((((------((((((-((----))))))))--)))))-))))-)))------------((-((-(((-------------------)))--))-))-----------((((((----((((-----(((------)))-----))))-))))))--)))))))-)))---))-)))))-----))))---))))-)))))(((-(----((((((---(((---))))))))))-)-))--)))))))))))))))----)))--)))-(((---)))-(((((((----)))))))---))))))))--))--))))-------------------------
//
Other, secondary structures (including those from Supplementary Figure S1 in [van.Nues-2007]):
# STOCKHOLM 1.0
#=GF SS From Suppl. Figure S1 https://doi.org/10.1016/j.jmb.2007.02.056
#=GS SacCas-AJ550804/1-549 DE AJ550804:101-633 Saccharomyces castellii
SacCas-AJ550804/1-549 AGGCTGTAATGGCTTTTAGTGGAACCAAGTATTTTGGGATATTTGGCCTGGGAACGAAACTTCCTTACGGTCAGACATGGATTGCACGTTTCGTCTTTGGCGGAACAAGGGGTTTTGCCAAGTTTTGGTAGGGTTCTGCGCCATGCCCTGTTGCTTCAACCGTCTTCCTGAACCCGCCTGTTAGGGCTTGTCTCGGCTGGTGTAGTTTTCTGTGCCAGTTGGAGGGTTTAGGGCTCTGAGTTAACGTGTCCCGGTTTAACAATCAATCGATCTTCGCGGGTTGCTCGTTGGCAGACAGCGCGAGAAATCCGTTGTTTCTGTCCGATGCGGCAATGTAATCTGGTCATGTGACCTCGCCTTGACCGCCATTAGGGGAGATTCATCCAGCGATGGCAAAGGCGACCCGTGAGAGAGGCGACTGGGATGGCACTGTTCTTAGGGTTAACAAGTACAAGTTCGGTTTATTATGGACGGTGGAGTCTGGTATTTCCTACCACATGTTGGGTGTAAACTACTCTTAGGAACCACTAACTACTTTATTTCTTTTTT
#=GC SS (((((.....)))))(((((((..((((((((((((((((((((((((.(((((......)))))...)))))))(((((..(((..((((((((...))))))))..(((((((((((((...))))))))))))))))))))).((....(((((.(((((((..((((((((((((....))))...((.((((((((((((....)))))))))))))))))))).))((((((((..((((((((((((((..............((((((((((((((((((((((((((((((.((....))))))))..))))).)))).)))))........((((....))))(((((((....((((((...(((......)))...)))))).)))))))))))))).)))...)))))))...)))).)))))))))))(((.((((.........)))).))).))))))))))))..)))))..))))(((.....)))....))).))..))).))..)))))))..................
#=GC BEAR-notation eeeeennnnneeeeeGGGGGGGKKBBCCCBBCCCDDDDCCCggggggg[eeeeeooooooeeeee333gggggggEEEEEKKCCC::hhhhhhhhlllhhhhhhhh::=============lll=============CCCEEEEE:BBMMMMEEEEE{GGGGGGG::BBFFFFFFddddmmmmdddd:::bb[============mmmm============bbFFFFFF}BBHHHHHHHHKKCCCDDDDGGGGGGGWWWWWWWWWWWWWWCCCGGGGGGGeeeeeddddeeeeeffffff[bbmmmmbbffffff22eeeee]dddd]eeeee::::::::ddddmmmmddddggggggg####ffffff"""cccooooooccc333ffffff]gggggggGGGGGGG}CCCZZZGGGGGGGZZZDDDD}CCCHHHHHHHHccc[ddddrrrrrrrrrdddd]ccc:GGGGGGGEEEEEYYBBCCCYYDDDDcccnnnnnccc::::CCC}BBYYCCC}BBYYGGGGGGG::::::::::::::::::
//
# STOCKHOLM 1.0
#=GF SS From Suppl. Figure S1 https://doi.org/10.1016/j.jmb.2007.02.056
#=GS CanGla-HG323640/1-504 DE HG323640.1 TPA: Candida glabrata SRP RNA for signal recognition particle RNA
CanGla-HG323640/1-504 AGGCTGTAATGGCTATTTAGTGGAATTGTTCTCCTTGGGAATTTTGGTCTGGGTTCGAACCCTACGGCCAGACGCGGAGGCACGCCTTTAATTATTACACCGTTTCGGTGGGGGCAAAGGAGCAAGCTGGGCTCGCCCAGCCTGTTCCTGCACCGTGACTTCCCGTGAACTGCAGTCGTCTCCCCCCTGTGGGGAAGGCGAGCCACCCAGGGACGTGTCTCGGTTCATCCAACCAATCGATCCTTGCGGGCGTTCGTGGCCCCTGCTGGCGCTGTGAGTAATCCAGTGTCCTCTAGGGCCGGCAACGTAGCTACGGTCTACCCTCCCTAGGAGGGGGGCCCACCTTCACCGCCGTTAGGGGAGTTTCATCCAGCGATGGCAAAGGTGACCCGTGATGGAGGCGGTCGGGATGGCACTGGTCCCGTGGTTACCTACCTGCAGTCGCGGTGGCATTATCCTGCCCTTCAATGGGCTTTGGGATTATCAAGACCACAATTTTTTTTT
#=GC SS .((((.....)))).....((((..(((.(.((((.(((((((((((((.((((....))))...)))))))((.((.(((((.(((((.....((.(((((...))))).))..))))).)...((((((....)))))).))))))))((((((((((.((.((((.......)))).(((((.....))))).))..((((((...((((((((((((((((...............((((((((((.(((..((((((.((..(((((((.((....))))))).))..))))))))..)))........(((((.((((((...)))))))))))((((((....((((.....(((......))).....)))).)))))).))))))).)))...)))))))...))))..)))))))))))..........))))))))))..)))..)))((((.....))))....))))..).)))..))))...........
#=GC BEAR-notation :ddddnnnnndddd:::::DDDDKKCCC{A{DDDD:CCCCCCggggggg[ddddmmmmdddd333gggggggBB{BB{DDDDa[eeeee$$$$$bb[eeeeellleeeee]bb22eeeee]a:::ffffffmmmmffffff:DDDDBBBBJJJJJJJJJJ:BB:ddddpppppppdddd:eeeeennnnneeeee:BB::FFFFFFLLLEEEEEDDDDGGGGGGGWWWWWWWWWWWWWWWCCCGGGGGGG:ccc!!ffffff[bb!!bbeeeee[bbmmmmbbeeeee]bb22bbffffff22ccc::::::::eeeee[fffffflllffffffeeeeeffffff####dddd$$$$$cccooooooccc55555dddd]ffffff:GGGGGGG}CCCZZZGGGGGGGZZZDDDDYYEEEEEFFFFFF@@@@@@@@@@JJJJJJJJJJYYCCCYYCCCddddnnnnndddd::::DDDDYYA}CCCYYDDDD:::::::::::
//
# STOCKHOLM 1.0
#=GF SS From Suppl. Figure S1 https://doi.org/10.1016/j.jmb.2007.02.056
#=GS SacKlu-AJ550805/1-453 DE (Saccharomyces) Lachancea kluyveri SRP RNA AJ550805:101..539
SacKlu-AJ550805/1-453 AGAGCTGTAATGGCTTTTGGTGGAACCAGGTGTCCCGAGAGTTCTGGTCTGGGTTCGAACCCTACGGCCAGACGTGGTTGCAATTATACAACTCTTTGTGGGTTGGCGCCGCGCCCAAGCTCAACTGCACTTCCTCGTAAGAGGAGCCGAGTTTTAGACGTGTCCCGGATTATATCGATCTTTGCGGGGAGTCCTGGGCGAGACATGAGAAGTCTGTCTTCGCTTAGGTACGGCAATGGTACCACACGACGCACCTTGCGTCGTTGTGTGGTTTTCGCATGCTCCTTCCGCTGAAGGGGAGTTTCATCCAGCACAGCAGGAGTCACCCGTGAGGAGGCGGCCGGGATGGCACTCGTCCTCGGTATCATTAAGCACGGCGGTGGGCTGGGATTTCCTCGGGGCATTTAGCCTCTGTTGGACACCTTAGGAACTACCAAAGTATTCTTTTTTTTT
#=GC SS_cons ((((((.....))))))((((((..((((((((((((((((((((((((.((((....))))...)))))))(((((.(((.......((((((.....))))))))))))))(((.(((((.(((((.((.((((....))))))(((((.....((((((((((((.((.......((((((((((..(((((((((((((((.((....))))))).)))))))).)).....(((.(((((((((((..........)))))))))))..)))..(((((((...((((....(((......)))....))))))))).))))))))))))...)))))))...))))..))))))))..............))))))))))))))))..))))((....))(((....)))))))))))..))..)))))).................
#=GC BEAR-notation ffffffnnnnnffffffFFFFFFKKBBHHHHHHHHDDDDCCCggggggg[ddddmmmmdddd333gggggggeeeee[ccc&&&&&&&ffffffnnnnnffffffccceeeeeCCC{EEEEE{EEEEE:bb[ddddmmmmddddbbEEEEENNNNNCCCDDDDEEEEE{BBPPPPPPPJJJJJJJJJJ::bbhhhhhhhheeeee[bbmmmmbbeeeee]hhhhhhhh]bb:::::ccc[===========ssssssssss===========22ccc::bbeeeee"""dddd####cccooooooccc4444ddddeeeee]bbJJJJJJJJJJZZZBBEEEEEZZZDDDDYYCCCEEEEE@@@@@@@@@@@@@@EEEEEEEEEECCCCCCYYDDDDbbmmmmbbcccmmmmcccHHHHHHHHYYBBYYFFFFFF:::::::::::::::::
//
# STOCKHOLM 1.0
#=GS KluWal-AADM01000319/1-418 DE AADM01000319.1:21425-21842 Lachancea waltii NCYC 2644 cont318, whole genome shotgun sequence
KluWal-AADM01000319/1-418 ATCACTGTAACGGTCGACAGTGGAACTGTATTTTCCGAGATTTCTGACCTGGGTTCAAACCCTGCGGTCGGACGTGTTACGCTTTAAGCCTCTTTCGGGAGGTTGCTACACGCTCAGTCTAACAATCGAGTCGACGTGCCCTGGTAACAAATCGATCCTTGCGGGCAGTCCAGGGCGAGATGTGAGAAGTCCGTCTTGTACCTGGTACGGCAATGGTAGGCGAGGGTGCTTAACGCTAACCCGTCTCTTGTGCTCCTATCGCTGAAGGGGAGTTTCATCCAGCACAGCAAGAGCAACCCGTGATGGAGGCGACTGGGATTACACTTGTCCTCGTATAAGGGATTTATCCCTTCGTTGTGGGCGTGAATCTCATCCTATGGAATGTGAAGATACTAGAACCCTGTCATATTATTTTTTA
#=GF SS Edited Suppl. Figure S1 https://doi.org/10.1016/j.jmb.2007.02.056
#=GC SS ...(((.....))).(((((.((..((((((((((.(((((((((((((.((((....))))...))))))).(((.((.((....(((((((....))))))))))))))(((((...((((...((((.(((.(((.((((((..........((((((((((..(((((((((((((((.((....)))))))))).))))).))....(((((((.((.((....))..))))).)).))......(((((.....((((....(((......)))....))))..))))).))))))).)))....))))))....))))))..))))...((((((....)))))).)))))))))...)))))).(((...)))....)))))))).))..))))))).............
#=GC BEAR-notation :::cccnnnnnccc:EEEEE{BBKKBBHHHHHHHH:FFFFFFggggggg[ddddmmmmdddd333ggggggg:ccc[bb[bb####gggggggmmmmgggggggbbbbcccEEEEELLLDDDD:::DDDD{CCC{CCC{FFFFFFSSSSSSSSSSCCCGGGGGGG::bbeeeeehhhhhhhh[bbmmmmbbhhhhhhhh]eeeee]bb::::bbbbccc[bb[bbmmmmbb22bbccc]bb]bb::::::eeeee$$$$$dddd####cccooooooccc4444dddd22eeeee:GGGGGGG}CCC~~~~FFFFFF~~~~CCCCCCYYDDDD:::ffffffmmmmffffff:DDDDEEEEEZZZFFFFFF:ccclllccc::::HHHHHHHH}BBYYBBEEEEE:::::::::::::
//
# STOCKHOLM 1.0
#=GF SS Edited domain 11 Suppl. Figure S1 https://doi.org/10.1016/j.jmb.2007.02.056
#=GS ZygRou-AJ564197/1-533 DE AJ564197 1-533 Zygosaccharomyces rouxii
ZygRou-AJ564197/1-533 AGCTGTAATGGCTCAATGGTGGAATTGTACACCTTGGGATTTCTGACTTTGGTTCAATCCTCGCAGTCGGACGTAGGTTAGCATGTCACGTTTCGGCGTGGCAAGGGGTACTCACCTTCCCGTGGGGCTCCGCACTGCGCGTCGGCGACAACCGTCATCCTTCTCTCGCTTCCAGTTGCCGAGGCCGTCAATCTTGGCGGTCTCAGAGAGAGGGACTCGGGGTTAACGTGTCCCGGTTATTATCGATCTTCGCGGGGTTTGTTTGCGCGTTCGGGCACGAGAGAAATCCGTGTTCCTGGGCGTGTGGCAAAGTAGTTACGGGTTCCTTGTGGGCCTCGCCTTGACCGCTGAAGGGGAGTTTCATCCAGCACAGCGAGGGCGACCCGTGATGGAGGCGGCCGGGATAGCACTAGTTCTCCGAGTACAAGTGCAGACTGGGTTGGCCCTGTGCCAATCTTAACGGGCGGTGTCGCTGATAATTACCCACCAGTAAAGGTACGTTGCTTGCAGCGTGGGGTGTACAAAACTATTTT
#=GC SS ((((.....))))....(((((..((((((((((.((((((((((((((.((......))..).)))))))((((.((..((.((((((((....)))))))).(((((.(((((......)))))))))))))))))).(((((((((.((((((..((((((((((((...)))....((((((((((....)))))))))).)))))))))((((((((..((((((((((((((........((((((((((.((((((.(((((((((((((((.((....))))))..)))))))))))))))))...((((((((...))))).))).(((((((...(((((....(((......)))....))))))))))))))))))).)))...)))))))...))))..)))))))))))...(((....)))((((((((.....)))))))).....))))))))))))))))))..)))(((......))).(((.(((...))))))))))))))))..)))))..
#=GC BEAR-notation ddddnnnnndddd::::EEEEEKKJJJJJJJJJJ:CCCCCCggggggga[bboooooobb22a]gggggggDDDD{BBKKBB:hhhhhhhhmmmmhhhhhhhh:eeeee[eeeeeooooooeeeeeeeeeeBBBBDDDD:IIIIIIIII{FFFFFF::IIIIIIIIIccclllccc::::==========mmmm==========:IIIIIIIIIHHHHHHHHKKCCCDDDDGGGGGGGQQQQQQQQCCCGGGGGGG:ffffff[===========dddd[bbmmmmbbdddd22===========ffffff:::ccceeeeellleeeee]ccc:ggggggg"""eeeee####cccooooooccc4444eeeeegggggggGGGGGGG}CCCZZZGGGGGGGZZZDDDDYYCCCHHHHHHHH:::cccmmmmccchhhhhhhhnnnnnhhhhhhhh:::::FFFFFFIIIIIIIIICCCYYCCCcccooooooccc:ccc[ccclllccccccJJJJJJJJJJYYEEEEE::
//
# STOCKHOLM 1.0
#=GS ZygBai-HG316458/1.525 DE HG316458.1:c717351-716827 Zygosaccharomyces bailii CLIB 213 genomic, scaffold05
ZygBai-HG316458/1.525 AGCTGTAATGGCTTTGGTAGAACTGTACGCTTCGAGATTTTCGGCTTGGGTTCAATCCTTACGGTCGGACGTAGGTTAGCGTGTCATGCTTCGGTGTGACAAGGGGTTCCTGTGAAAGCAGGAATTCTGCACTGCGCGTCGGTGCCAACCACTATCCTCTCCCTGTTTCCATTTGCGGAGGCGACTAGCCTATTAGGTTACCTCTGGGGGAGGACCTGGAGTTAACGTGTCCCGGTACTTATCGATCTTTGCGGGTTTGCAGAGCCTAGGCACGAGAGAAGTCCGTGATCTAGCCTGTGGCAACGTAGTATGGGCTCTTTGTAGCCTCGTCCTCACAGCTGTAGGGGAGATTCATCCAGCACAGCAGGGGCGACCCGTGACGGAGGCGGTCGGGATAGCACTAGTTCTCCGGGTATTAGTGCAGTGTGGGCTCGCTTGTCGGGTAACATTTGGTGGTGGCACTGATAATTGCTCCTTTGTTAAGCCTCTGGTTCTCCAGTGGGGAGCGTATTGAACTACTTTTTT
#=GC SS ((((.....))))..(((((..(.((((((((((((((((((((((.(((......)))...)))))))((((.((..((.((((((((....)))))))).(((((((((((....))))))))))))))))))).(((((((((.((((((..((((((((((....)).....(((((.((((.........)))).)))))))))))))((((((((..((((((((((((((........((((((((((((((.(.((.(((((((((.((....)))))).))))).)).).))))....(((((((...))))).)).(((((((...(((((....(((......)))....))))))))))))))))))).)))...)))))))...))))..)))))))))))........(((((..(((((.....))))).))))).))))))))))))))))))..)))(((....)))((.((((....)))).))))))))))).)..))))).....
#=GC BEAR-notation ddddnnnnndddd::EEEEEKKA{IIIIIIIIICCCCCCggggggg[cccooooooccc333gggggggDDDD{BBKKBB:hhhhhhhhmmmmhhhhhhhh:===========mmmm===========BBBBDDDD:IIIIIIIII{FFFFFF::HHHHHHHHbbmmmmbb:::::eeeee[ddddrrrrrrrrrdddd]eeeeeHHHHHHHHHHHHHHHHKKCCCDDDDGGGGGGGQQQQQQQQCCCGGGGGGGdddd[a[bb[eeeeedddd[bbmmmmbbdddd]eeeee]bb]a]dddd::::bbeeeeellleeeee]bb:ggggggg"""eeeee####cccooooooccc4444eeeeegggggggGGGGGGG}CCCZZZGGGGGGGZZZDDDDYYCCCHHHHHHHH::::::::eeeee!!eeeeennnnneeeee]eeeee:FFFFFFIIIIIIIIICCCYYCCCcccmmmmcccbb[ddddmmmmdddd]bbIIIIIIIII}AYYEEEEE:::::
//
# STOCKHOLM 1.0
#=GF SS From Suppl. Figure S1 https://doi.org/10.1016/j.jmb.2007.02.056
#=GS EreGos-HG323639/1-381 DE HG323639 Ashbya gossypii ATCC 10895 SRP RNA for signal recognition
EreGos-HG323639/1-381 AGCTGTGACGGCCTTTGGCCGGAAGGGACATTAGCAATCCCGGCTTGGTGAGGCGCTCCCGCGTTCGAACCTGCGGCCGGGCATGGTTGCGGAAAAGCGAGGTTGCCTAGCTCCTCGTGCGCCATGATACCACACTGGTCTTTCTGACGCGTGTCCTGCTACTTATCGATTTCTGCGGGTTTGCCGTCGATGCTGACATGGGAAACCTGTCGGTGGGGCGTGCAGTAATGGCAGGGCGAATGCCCGTCCGCTCATACGCTGGAGGGGAGTTTCATCCAGCACAGCGCGAGCGGCCCGTGGGGAGGCGAGCAGGATGGCACGTGTTAGGCGCTTGCGCGGACCTATCGTGGTATTGGAGCATTATGTTCCGACCGACTTTTT
#=GC SS -(((.....)))----((-(((--(((((((-(-(((-(((((((-(((--(((((----)))-))--)))---)))))))(((((-(((------((((((--((---))-))))))))))))))-((((((---(((((--(((((((((((((((((------...((((((((((-((((((((--((((((((-((.-.-))))))))))-))))-))))--((((((-------)))-)))((((((---(((((-...(((.----.)))...-)))))-))))))))))))))))...-))))))---)))))))))))(((----))))))))----)))))))))-----)-)))))))--)))-))----
#=GC BEAR-notation :cccnnnnnccc::::BB{CCCKKGGGGGGG{A{CCC:ggggggg[ccc!!bbcccmmmmccc]bb22ccc333gggggggeeeee[ccc%%%%%%ffffff!!bblllbb]ffffffccceeeee:FFFFFFLLLEEEEE::JJJJJJJJJJJFFFFFFRRRRRRRRRJJJJJJJJJJ:dddddddd!!hhhhhhhh[bbmmmmbbhhhhhhhh]dddd]dddd::ccccccpppppppccc]cccffffff"""eeeee####cccooooooccc4444eeeee]ffffffJJJJJJJJJJ~~~~FFFFFFZZZJJJJJJJJJJJcccmmmmcccEEEEE~~~~FFFFFFCCC?????A}GGGGGGGYYCCC}BB::::
//
# STOCKHOLM 1.0
#=GS KluDob-PPJP02000011/1-523 DE PPJP02000011.1:c238726-238204 Kluyveromyces dobzhanskii strain NRRL Y-1974 NODE_11_length_285983_cov_19.1963_ID_21, whole genome shotgun sequence
KluDob-PPJP02000011/1-523 ACAGCATCCTGGCGAAGGAGATAGCATCTAGGACATGTGCCTCCGGTTCGAAACCGCACAGGCACACCGGGTCCCTTTCAATGAGAGAGGTCTACGGATGGTACCCCAAACAGGGGCGGCGCTGTCACGTGTCCATTGGTCTAACCGCCAATTCCGCGTGCTGCACCGTTCTTGGCCGCCCGGTCTACGCGTTCATAGCTTCGTGCTTCCAACTGTTTTGGACAGCGCCATCCTGGCGCTGTCACGTGGTCGACGTGTCCCATCGTTTGAAAATTGTTGCGGGTTATCTTTGTGGTGGCTCGGTCACGTGAGAAGTCCGTGATTTGAAGTCCAACTGGTAACGATTTAGCTTCAACATCGGTCCAATTTCATTGGATTAGAAGCTAGTTCAGTGTTCATCGTTGTAGGGGAGTTTCATCCAGCACAACAAACACTGCCCGTGACGAGGTGGTGCGGATTGCACGTCCCATGGTAAAGCAGTACGTGTGGGTTCACAGGTTGCTTACAGCCCTTCATATTTTTT
#=GC SS_cons .(((....)))..(.(((.(.(((((.(((((((.((((((..(((((...)))))....))))))((((((.((((((.....))))))((....)).(((........((((((((((((...(((((...(((((.((((....))))))))).)))))))))))))))))))))))))).((((((((....(((.....)))....((((((((.((((((((((...))))))))))((((((..((((((((((((((((......(((((((((((((((((.((..((((..(((((((.((....))))))))).)....))))).)))))....((((((((......((((((((...)))))))).))))))))..(((((((....(((((...(((......)))...))))).)))))))))))))))))))))))).))...))))))))))).))))))))))))))))))))))..))).))).)))...))))).).......
#=GC BEAR-notation :cccmmmmccc::A{CCC{A{ACCCA{BBCCCBB:ffffff!!eeeeellleeeee4444ffffffFFFFFF:ffffffnnnnnffffffbbmmmmbb:ccc''''''''============"""eeeee"""eeeee[ddddmmmmddddeeeee]eeeee============cccFFFFFF:HHHHHHHH::::cccnnnnnccc::::HHHHHHHH:==========lll==========DDDDBBKKIIIIIIIIIBBEEEEEOOOOOOJJJJJJJJJJJJeeeee[bb!!ccca!!ggggggg[bbmmmmbbggggggg]a4444cccbb]eeeee::::hhhhhhhh%%%%%%hhhhhhhhlllhhhhhhhh]hhhhhhhh::ggggggg####eeeee"""cccooooooccc333eeeee]gggggggJJJJJJJJJJJJEEEEE}BBZZZIIIIIIIIIBB}DDDDHHHHHHHHHHHHHHHHBBYYCCC}BBA}CCCZZZAACCC}A:::::::
//
# STOCKHOLM 1.0
#=GS KluWic-AEAV01000057/1-440 DE AEAV01000057.1:40798-41237 Kluyveromyces wickerhamii UCD 54-210 contig00060, whole genome shotgun sequence
KluWic.AEAV01000057/1-440 ACCTGTCGTGGTACAAGGGATGTGATCAGTATAAGATATGTGGCTCCGGTTCGAAACCGCACAGCTGCACTGGGTCCGGTATTCCAAACAGCAACTTTCTTTCGGGAATTCTTGTTGGCTGCCCAGCTTCTGCGGAATCAAAACCACTTTGGCTACAATACGTGGTCTCGTGGTTGACGTGTCCCGTTGTTTCAAAATCGTTGCGGGTTTATTAATGAGGCTCCACATGAGAAATCTGTGCAAAGCGTCTATTTGGTAATTGTTCCGCATTCAGGTTCTATGCCTGAATCGGTCACGTGTTTACCGTTGGAGGGGAGTTTCATCCATCACAACAAACACGGCCCGTGACGAGGTAGCGTGGATTGCATGTCCCATGTCCTAAGTGGTACGCGGAAAATCTTGTAATTCAGGCTGGACACAAACCCTTGGCTTCTTTTTTC
#=GC SS_cons (((......))).((((((.((((.((((((((((((.(((((((.(((((...)))))...)))))))((((((((((((........((.((.(((((...)))))))))))))))..)))))).(((((((........(((((((.((((((.....)))))).(((((..((((((((((((((((......((((((((((.(((((((..((((..((((.((....))))))......))))..))))))).((..(((.((((((((.....))))))))))).))((((((....((((....(((......)))....)))).)))))).))))))))))))))))).))...))))))))))))....))))))).))))))).))))))).)))..)).....))))..))))))............
#=GC BEAR-notation ecccooooooccc:FFFFFF{DDDD{BBCCCGGGGGGG:ggggggg[eeeeellleeeee333gggggggffffffffffff''''''''bb[bb[eeeeellleeeeebbbbffffff22ffffff:GGGGGGGQQQQQQQQGGGGGGG:ffffffnnnnnffffff:EEEEEKKGGGGGGGBBGGGGGGGOOOOOOJJJJJJJJJJ:ggggggg!!dddd!!dddd[bbmmmmbbdddd666666dddd22ggggggg:bb!!ccc[hhhhhhhhnnnnnhhhhhhhhccc]bbffffff####dddd####cccooooooccc4444dddd]ffffff:JJJJJJJJJJGGGGGGG}BBZZZGGGGGGGEEEEE~~~~GGGGGGG}GGGGGGG}GGGGGGG}CCCYYBB?????DDDDYYFFFFFF::::::::::::
//
# STOCKHOLM 1.0
#=GS KluNon-QYLQ01000002/1-421 DE QYLQ01000002.1:867865-868285 Kluyveromyces nonfermentans strain NRRL Y-27343 flattened_line_2, whole genome shotgun sequence
KluNon-QYLQ01000002/1-421 ACTGTTGTGGCATGGTAAAGCGGAAGCGAGCACTTTGAAGATGTGTCCCCGGTTCAAAACCGTACGGTCACAGTGGCATGGTAATACAAACAATAACACACATTTGTGTGTTATTGGCCACCCACTGCTCAAGTAACCATCTCAGTTTGTTATTGTTCAAGCTGCGCGGTTGACGTGTCCCATTGTTTCAAAATCGTTGCGGGTGTCTAAGTCTGGTCACGTGTAAGTAATTCGCGTTGATCAGCTGTTTGGCAATTGTAGTAGGTTTTCTACGCACGCGTGAACAGTTGGAGGGGAGTTTCATCCAGCACAACGAGCGCGGCCCGTGATGAGGTGGTGAGGATTGCACGTCCCGCGAAATGATGGTACTGGGTTCTTCAGCACGGTGTTGTTACATACCGCATTTCTCTTTTTCCTTTTT
#=GC SS_cons (((((......)))))...((((((.(((.(((((((((((((.(((..(((((...)))))...)))))((((((..((((........(((((((((((....))))))))))))))).))))))(((.(((.((((((..((((((........)))))).(((((..((((((((((((((((......((((((((((((.(((((((((((((((((.((....))))))).))))))..))))))))..(((.((.((....)))))))((((((....((((....(((......)))...))))).)))))).))))))))))))))))).))...))))))))))))....)))))))))))).)))).))...))))))))).....))))...................
#=GC BEAR-notation eeeeeooooooeeeee:::DDDDBB{CCC{DDDDBBDDDDAbb[ccc!!eeeeellleeeee333cccbbffffff!!dddd''''''''===========mmmm===========dddd]ffffffCCC{CCC{EEEEEA::ffffffqqqqqqqqffffff:EEEEEKKFFFFFFBBHHHHHHHHOOOOOOIIIIIIIIIAbb[ffffffffffffeeeee[bbmmmmbbeeeee]ffffff22ffffffbb::ccc[bb[bbmmmmbbbbccceeeeea####dddd####cccooooooccc333dddda]eeeeeA}IIIIIIIIIHHHHHHHH}BBZZZFFFFFFEEEEEA~~~~EEEEECCCCCCA}DDDD}BBZZZDDDDCCCBB?????DDDD:::::::::::::::::::
//
# STOCKHOLM 1.0
#=GS KluAes-PPJO01000006/1-416 DE PPJO01000006.1:54130-54545 Kluyveromyces aestuarii strain NRRL YB-4510 NODE_6_length_597287_cov_27.2088_ID_11, whole genome shotgun sequence
KluAes-PPJO01000006/1-416 ACATCTGTACTGGTTTGACGGACTGGAACGGCTTAGGATACGCGACCCCGGTTCGAAACCGCACGGTCGTGATGGGTCGGTAACTCAAACAAAAACTCACGTCTCGTGTTTCTTTGGCCGCCCATGCTTCTGTTAGCCACCTTGACCCAGGCATTTAGCTTTGGTCACGTGGTAGACGTGTCCCATTGTTTCAAAATCGTTGCGGGTGTCTATGGCTGGTCTCGTGTGAGTAGTCCGCGTTGGGCAGTCGTTTGGCAATTGCAATAGGTTATCCCTATGTTACACGTGAACAGTTGGAGGGGAGTATAATCCAGCACAACGAGCGTGGCCCGTGATGAGGTGGTGCGGACTGCACGTCCTACGATATAGGTGGTACAGTTGTTGTTCTGTCATGTTGGTCTAAACCGTCTTTTTTT
#=GC SS_cons ((....))..(((((((((.(((((........((((((.(((((((.(((((...)))))...)))))))((((((.(((........((((((..((((...)))))))..))).))))))))((..((((..((((((((((.((((((.....))).))))))(((((..((((((((((((((((......((((((((((((((.(((((((.(((((((.((....))))))..))))))))))..))))...((.(((((.....)))))))..((((((....((((....(((......)))....)))).)))))).))))))))))))))))).))...))))))))))))....)))))))))))..))))))))).)).))).)).))))))).........
#=GC BEAR-notation bbmmmmbb::GGGGGGGBB{CCCBBQQQQQQQQFFFFFF:ggggggg[eeeeellleeeee333gggggggAeeeee[ccc''''''''cccccc!!ddddlllddddccc22ccc]ccceeeeeBBKKDDDDKKGGGGGGGccc[ccccccnnnnnccc]ccccccEEEEEKKGGGGGGGBBGGGGGGGOOOOOOJJJJJJJJJJdddd[ggggggg[cccdddd[bbmmmmbbdddd22cccggggggg22dddd:::bb[eeeeennnnneeeeebb::ffffff####dddd####cccooooooccc4444dddd]ffffff:JJJJJJJJJJGGGGGGG}BBZZZGGGGGGGEEEEE~~~~GGGGGGGDDDDYYBBAFFFFFF}BB}CCC}BB}GGGGGGG:::::::::
//