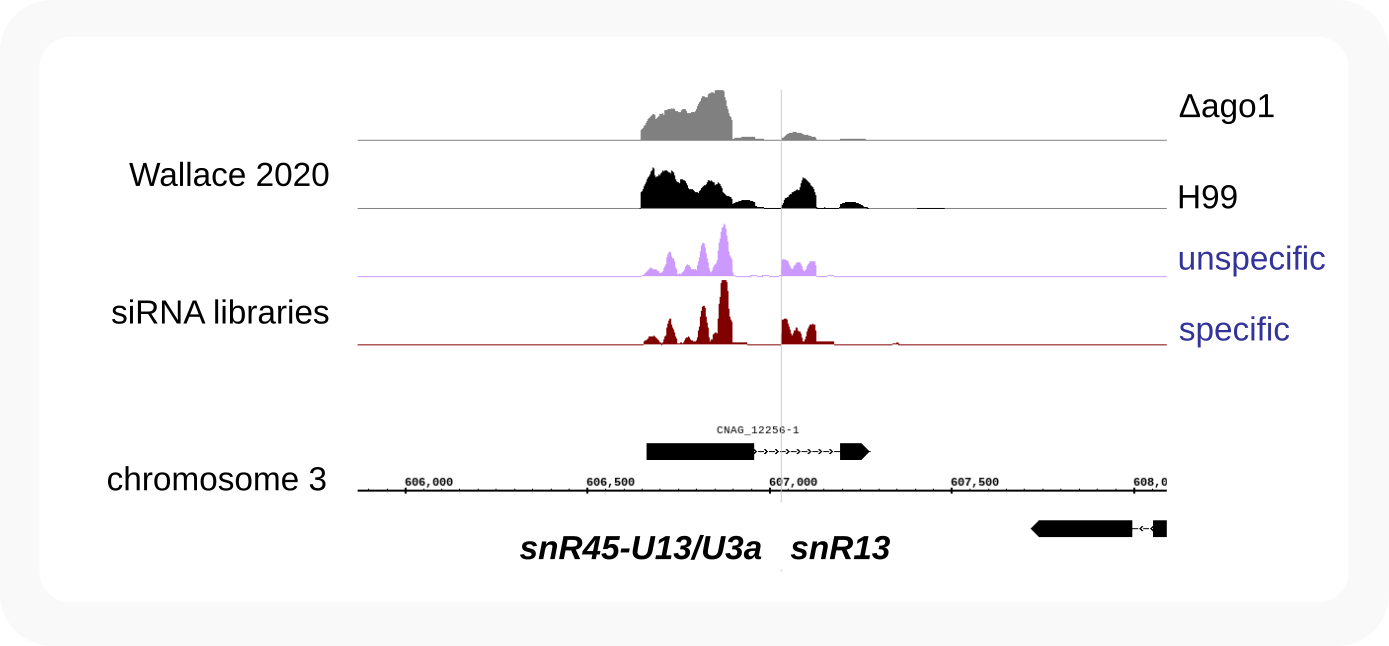
CNAG_12256¶
Non-coding transcript (CNAG_12256) with snoRNA snR45-U13_U3a on the first exon and snoRNA snR13 within the spliced intron.
snR13¶
snR13_intronic-boxCDsnoRNA
Within transcript as CNAG_12256
Multiple modification by snR13 relates to the C linking the guide to the D’ box (ttga) in
5'-TGAGtGCATTTGGCTTGA-3'. This C can become the edge of the D’ box that defines modification by Nop1/fibrillarin at the 5th nucleotide in the target base paired to the guide. In this alternative configuration the G in the target sequence might remain base-paired via the accessory guide [van.Nues-2011](gctacctt) that anchors the aaggta upstream of the target sequence. Without this additional base-pairing potential the alternative modification by snR13 does not occur (see [van.Nues-2016]).Orthologue of yeast snR13, for D'-box guide: human SNORD15A or SNORD15B (D-box)

>AE017343.1:c1423298-1423205 Cryptococcus neoformans var. neoformans JEC21 chromosome 3 sequence ACCGAACGATGACCTGAGTGCATTTGGCTTGAGGACGTCTTTACCGTCGTCGATGCTGAAAAAACCACGC TACCTTACTTTTTACCTCTGACAA
snR45-U13_U3a¶
Processed from a transcript that gets spliced (as CNAG_12256)
snR45-U13_U3a_exonic-boxCDsnoRNA-253nt
Almost the same sequence as U3b snoRNA on chromosome 8
The homologue from Naematelia encephala (URS0000D98D81_71784 ) is assigned by Rfam as U3 (Rfam RF00012)
The 5’ stem-loop is in Tremellomycetes relatively short, while this structural element is quite extensive in yeast U3 with complimentarity to ETS1 sequences and those forming the pseudo-knot in 18S rRNA.
In baker’s yeast, apart from U3, snoRNAs snR4 and snR45 contain a leader sequence with base-pairing functionality [Sharma-2017].
snR45/U13 and snR4 associate with Kre33/NAT10 (CNN01050, CNAG_06379) to guide this nucleobase acetyltransferase to the target cytosine which gets acetylated in a mechanism similar to pseudouridylation supported by H/ACA snoRNAs; Kre33/NAT10 also modify a group of tRNAs (see [Sharma-2017]).
If the same cytosines in Cryptococcus are modified as in baker’s yeast, this U3-like snoRNA might guide Kre33/NAT10 like snR45.
Predicted target in 18S rRNA: [gtaacaa]ggtttcCgtaggtgamamcctg[c]g[g]a[a]ggatc[a]gta [2];
Orthologue of yeast snR45, U13 from human: SNORD13, or plant: SnoR105, SnoR108. Alternatively, this is one of two U3 snoRNAs in Cryptococcus, which in yeast is snR17a or snR17b, in human SNORD3@
Alignments:
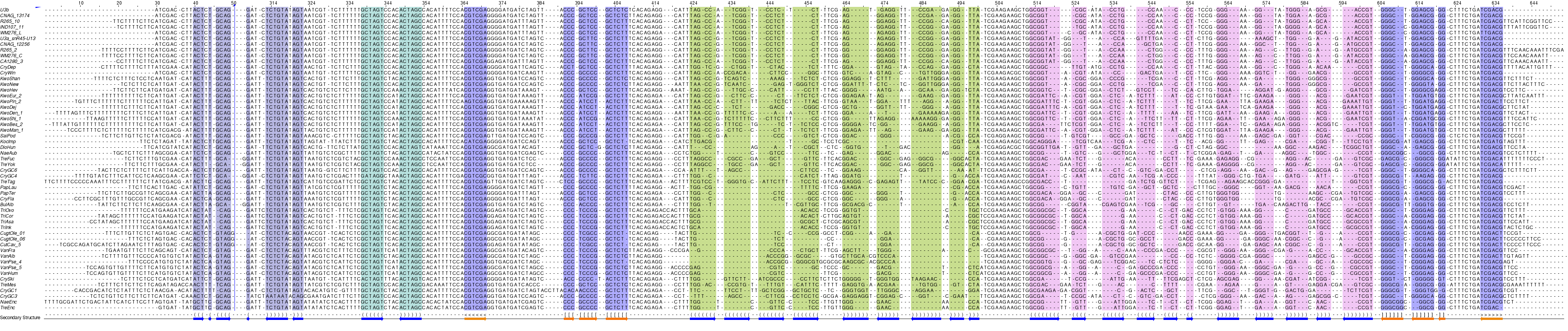
snR45/U3 homologues in Tremellomycetes¶
Putative secondary structure and interaction with 18S rRNA 3’ end:
Tremellomycetes snR45 U13 (mammal, plant)
Insets: proposed base pairing between the 3’ region of 18S rRNA and yeast snR45 or human U13 (from [Sharma-2017])
Another snoRNA (snR45-II) might provide or regulate base-pairing with the 5’ section of the target region near the 3’ end of 18S rRNA. The potential for that interaction seems more evident for that snoRNA than for snR45-U13.
The functions of yeast snR45 and U3 could have been divided over several snoRNAs in Tremellomycetes.
UV crosslinking experiments to isolate RNAs that bind specificly to Kre33/Nat10 from Cryptococcus (as done for yeast) might yield relevant information if U3 snoRNA fragments are not found as a common contaminant, which was the case for such experiments in baker’s yeast [Sharma-2017].
>AE017343.1:c1423686-1423434 Cryptococcus neoformans var. neoformans JEC21 chromosome 3 sequence ATCGACCTTACTCTGCAGGATCTCTGTATAGTAATCGTTCTTTTTTGCTAGTCCACACTAGCCACATTTTCC ACGTCGAGGGGATGATCTAGTCACCCGCTTCGCTCTTTCACAGAGGCATTTAGCCGTCGGTCCTCTCTTT CGAGAGAGGTTCCGGGAGGTTATCGAAGAAGCTGGCGGTATGGTACCAGTTTTGACCTCTTGGGGAAAGC TTGGGAGATACCGTGGGCTGGAGCGGGCTTTCTGATCGACG
Notes¶
Image source snR13:
# STOCKHOLM 1.0
#=GF RW van Nues, May 2023
#=GF https://coalispr.codeberg.page/paper
Annot/1-43 ---------------------RTGATGA---GCUC-CGUAAACCGCTGA------------------------------------------------------------------------RTGATGA---------CGAUGGAA-------------CTGA----------
#=GS intronic-snR13/1-101 DE ncrna 3:1423203:1423300:-1
intronic-snR13/1-101 --------------TTACCGAACGATGACC-TGAGTGCATTTGGCTTGAGGACGT---C-----------------------TTTACC-----------------------G--T-CGTCGATGCTGAAAAAACCACGCTACCTT--ACTTTTTACCTCTGACAAATTTT--
#=GS R265/1-98 DE CP025759.1:c1617388-1617291 Cryptococcus gattii VGII R265 chromosome 1, complete sequence
R265/1-98 ---------------TTACGAACGATGACC-TGAGTGCATTTGGCTTGAGGACGT---C-----------------------TTTACC-----------------------G--T-CGTCGATGCTGAAAAAACCACGCTACCTT--ACTTTTTACCTCTGACAATTT----
#=GS H99/1-99 DE CP003822.1:607033-607131 Cryptococcus neoformans var. grubii H99 chromosome 3, complete sequence
H99/1-99 --------------TTATCGAACGATGACC-TGAGTGCATTTGGCTTGAGGACGT---C-----------------------TTTACC-----------------------G--T-CGTCGATGCTGAAAAAACCACGCTACCTT--ACTTTTTACCTCTGACAATTT----
#=GS WM276/1-98 DE CP000288.1:c1378894-1378797 Cryptococcus gattii WM276 chromosome C, complete sequence
WM276/1-98 ---------------TTACGAACGATGACC-CGAGTGCATTTGGCTTGAGGACGT---C-----------------------TTTACC-----------------------G--T-CGTCGATGCTGAAAAAACCACGCTACCTT--ACTTTTTACCTCTGACAATTT----
#=GS KwoHev/1-97 DE ASQC01000169.1:c6788-6692 Kwoniella heveanensis CBS 569 cont2.169, whole genome shotgun sequence
KwoHev/1-97 -----------TCCCATTTGAACGATGACC-TGAGTGCATTTGGCTTGAGGACGT---C-----------------------TCTCGC-----------------------G--T-CGTCGATGCTGAAAAAACCACGCTACCTT--AAACTTTACCTCTGAC---------
#=GS CryWin/1-102 DE CP034262.1:307503-307604 Cryptococcus wingfieldii strain CBS7118 chromosome 2, complete sequence
CryWin/1-102 ------------TCACCACGAATGATGACC-CGAGTGCATTTGGCTTGAGGAC-T--GCT----------------------TTACT-----------------------AGT---CGTCGATGCTGAAAAAACCACGCTACCTT--AAATTCAATCTCTGACAACTT----
#=GS KwoDej/1-105 DE ASCJ01000004.1:1657359-1657463 Kwoniella dejecticola CBS 10117 cont1.4, whole genome shotgun sequence
KwoDej/1-105 ------------------TCTATGATGACC-TGAGTGCATTTGGCTTGAGGAC-T---C--------G--TGGA--TCC---TTTTT----GGG--TCCA--G--------G--TC-GTCGATGCTGAAAAAACCACGCTACCTT--AAATTCAATTTCTGACA--------
#=GS KwoBes/1-115 DE ASCK01000010.1:c185588-185474 Kwoniella bestiolae CBS 10118 cont1.10, whole genome shotgun sequence
KwoBes/1-115 -------------TCTATCGATTGATGACC-TGAGTGCATTTGGCTTGAGGAC-T---CT----------TGAA--TCC---TTTTT----GGG--TTCA----------GG--TC-GTCGATGCTGAAAAAACCACGCTACCTT-AAACTTTTACCTCTGACAATTT----
#=GS KwoMan/1-114 DE ASQF01000028.1:c357617-357504 Kwoniella mangroviensis CBS 8886 cont1.28, whole genome shotgun sequence
KwoMan/1-114 -----------TTACTATCGATTGATGACC-TGAGTGCATTTGGCTTGAGGAC-T---CT----------TGAA--TCC---TTTTT----GGG--TTCA----------GG--TC-GTCGATGCTGAAAAAACCACGCTACCTT-AAACTTTTACTTCTGACAA-------
#=GS BulAlb/1-111 DE CAMYTR010000032.1:c72457-72347 Bullera alba genome assembly, contig: jcf7180000012125, whole genome shotgun sequence
BulAlb/1-111 ----------------TCGACTTGATGACC-CGAGTGCATTTGGCTTGATGG------C---------AGGAGT--TCC---CTTCTT---GGG--GCTCTT-C-------G---C--CCGATGCTGAAAAAACCACGCTACCTT-AAATTCAATTTTCTGATACCT-----
#=GS CutCya/1-109 DE BEDZ01000044.1:98135-98243 Cutaneotrichosporon cyanovorans DNA, scaffold: scaffold_44, strain: JCM_31833, whole genome shotgun sequence
CutCya/1-109 ---------------CCATTCCGGATGACC-CGAGTGCATTTGGCTTTAGGG--T-CGCG-------C------AACACC---TT-----GGTG-G------AC------CGCG-C--CCACTGCTGAAAAACCCACGCTACCTT-AACTTAAACTTTCTGAAACC------
#=GS CutDas/1-110 DE BEDX01000002.1:755160-755269 Cutaneotrichosporon daszewskae DNA, scaffold: scaffold_2, strain: JCM_11166, whole genome shotgun sequence
CutDas/1-110 ---------------CCTTTCACGATGACC-CGAGTGCATTTGGCTTTAGGG--T---CG---------CGCGG--CGCA---TTC----TGTG--TCGCG---------TG---C--CCACTGCTGAAAAACCCACGCTACCTT-AAACTTTTTCTTCTGACACC------
#=GS CutOle/1-132 DE JZUH01000053.1:c22286-22155 Cutaneotrichosporon oleaginosum strain IBC0246 CC85scaffold_27_Cont53, whole genome shotgun sequence
CutOle/1-132 ---------------CAGTACTTGATGACC-CGAGTGCATTTGGCTTTAGGG--A-CACTGTGCCC-C-CTCGT-TCCCACC-TT---GGTGGG--GCGGG-ATGGGCCCGGTG-G--CCTCTGCTGAAAAACCCACGCTACCT--AAACAAACTTTTCTGAAACC------
#=GS CryCur/1-108 DE BCJH01000009.1:c565427-565320 Cryptococcus curvatus DNA, scaffold: scaffold_8, strain: JCM 1532, whole genome shotgun sequence
CryCur/1-108 ---------------CCTTTCCTGATGACC-CGAGTGCATTTGGCTTTAGGG--T------------C-GCGCG-ACACT---TT-----GGTGAA-C------CGTG-C--------CCACTGCTGAAAAACCCACGCTACCTTT-AATTCAACTTTCTGACAC-------
#=GS PapTer/1-106 DE JAHXHD010000715.1:c2761-2656 Papiliotrema terrestris strain LS28 scaffold-714, whole genome shotgun sequence
PapTer/1-106 -------------CTGTTTGCACGATGACC-CGAGTGCATTTGGCTTGAGGGC------G--GC------TGGT---------TC-----------GCCA-----GC---T---TA-GCCGCTGCCGAAAAACCCACGCTACCTT--AACTTAAACTTCTGAAAATCT----
#=GS TriAsa/1-113 DE BCLT01000004.1:c1206616-1206504 Trichosporon asahii DNA, scaffold: scaffold_4, strain: JCM 2466, whole genome shotgun sequence
TriAsa/1-113 --------------CGCTTGAATGATGACC-AGAGTGCATTTGGCTTTAGGG--C---CG--GCG-AC----TA--CCA----TTC-----TGG--TA-----CCGC---TG--T---CCTCTGCTGAAAAACCCACGCTACCTT--AACTTAAACTTCTGACGAACC----
#=GS TriFae/1-115 DE JXYK01000003.1:1200405-1200519 Trichosporon faecale strain JCM 2941 scaffold_0003, whole genome shotgun sequence
TriFae/1-115 ----------------CTTGAATGATGACC-AGAGTGCATTTGGCTTTAGGG--C---CG--GCG-AC----TG--CCA----TTC-----TGG--CA-----CCGC---TG--T---CCTCTGCTGAAAAACCCACGCTACCTT--AACTTAAACTTCTGACGAACACCTT
#=GS SaiPod/1-91 DE CABVSH010000546.1:c29750-29660 Saitozyma podzolica genome assembly, contig: NODE_549_length_31319_cov_13.8927, whole genome shotgun sequence
SaiPod/1-91 ---------------CCTTTGCGGATGACC-AGAGTGCATTTGGCTTGAGGACG----CC-----------------------TC-------------------------GG----TGTCGCTGCTGAAAAAACCTCGCTACCTT--ACTATTTCTTTCTGAAAC-------
#=GS ApiDom/1-107 DE BCFW01000004.1:c2563194-2563088 Apiotrichum domesticum DNA, scaffold: scaffold_3, strain: JCM 9580, whole genome shotgun sequence
ApiDom/1-107 -----------------CTACATGATGACC-CGAGTGCATTTGGCTTTAGGG--GA--CTG-----GC----GC--GCC---GTTA-----GGC-AGT-----------CGG--CA--CCTCTGCTGAAAAACCCTCGCTACCTT-AATAAACTCTTTCTGAAAC-------
#=GS TreFuc/1-94 DE BRDD01000065.1:c263583-263490 Tremella fuciformis NBRC 9317 DNA, KCNB80TF.65, whole genome shotgun sequence
TreFuc/1-94 ------------CTATCTCGATTGATGACC-CGAGTGCATTTGGCTTGAGGGTC----C------------------------TT--------------------------G----GACCGCTGCTGAAAAACCCACGCTACCTTT--AAATTTTTGTCTGACAACC-----
SacCer-snR13/1-124 AGGAAGTTTTTTCCTTTTTATATGATGAATATGAGTGCATTTGGCTCGA-GTTG----CT----------GTTT--GGC---TTTT-----GCC--AAAT---C------AG----TAACGGTGTGGAAAAACTCAAGCTACCTTTTTTTACTTTTATCTGACC--------
#=GC SS_cons --------------------------------------------------((((..(((((.((((..((((((..((((((......))))))..))))))..)))).)))))..))))----------------------------------------------------
//
Image source snR45-U13_U3a:
# STOCKHOLM 1.0
#=GF RW van Nues, May 2023
#=GF https://coalispr.codeberg.page/paper
#=GF R2R keep allpairs
#=GS Annot/1-22 DE boxCD elements; remove Annot lines before creating r2r figure.
Annot/1-22 ---------------------------------------------------------------------------------------------RTGATGA----------------------------CTGA----------------------------------------------------------------------RTGATGA----------------------------------------------------------------------------------------------------------------------------CTGA------
#=GS B3501/1-251 DE AAEY01000013.1:650841-651091 Cryptococcus neoformans var. neoformans B-3501A chromosome 3 chr03.040506.C2, whole genome shotgun sequence
B3501/1-251 ATCGACCTTA----------CTCTGCAGGATC-TCTGTATAGTAATCGTTCT--TTTTTGCTAGTCCACACTAGCCACATTTTCCACGTCGAGGGGATGATCTAGTCACCCGCTTC---GCTCTTTCACAGAGGCATTTAGCCG-TCGG--TC--CTC-TCTT-----TC-----GGGA--GAG--GTT--CCGGGAGGTTATCGAAGAAGCTGGCGGT--AT--------GGTACC---AGTT-GA---------CCTC--TT--GGGG----------AA--AGCTTGGGAG---------AT--ACCGTGGGC---TGGAGCG-GGCTTTCTGATCGACG
#=GS H99/1-251 DE NPNB01000012.1:606650-606900 Cryptococcus neoformans var. grubii strain H99 Chr_3, whole genome shotgun sequence
H99/1-251 ATCGACCTTA----------CTCTGCAGGATC-TCTGTATAGTAATCGTTCT--TTTTTGCTAGTCCACACTAGCCACATTTTCCACGTCGAGGGGATGATCTAGTCACCCGCTTC---GCTCTTTCACAGAGGCATTTAGCCG-TCGG--TC--CTC-TCTT-----TC-----GGGA--GAG--GTT--CCGGGAGGTTATCGAAGAAGCTGGCGGT--AT--------GGTACC---AGCT-GA---------CCTT--TC--GGGG----------AA--GGCTTGGGAG---------AT--GCCGTGGGC---TGGAGCG-GGCTTTCTGATCGACG
#=GS R265/1-251 DE NPNA01000004.1:589903-590153 Cryptococcus gattii VGII strain R265 tig00000004, whole genome shotgun sequence
R265/1-251 ATCGACCTTA----------CTCTGCAGGATC-TCTGTATAGTAATCGTTCT--TTTTTGCTAGTCCACACTAGCCACATTTTCCACGTCGAGGGGATGATCTAGTCACCCGCTTC---GCTCTTTCACAGAGGCATTTAGCCG-TCGG--TC--CTC-TCTT-----TC-----GGGG--GAG--GTT--CCGGGAGGTTATCGAAGAAGCTGGCGGT--AT--------GGTACC---AACT-GG---------CCCT--TT--GGGG----------AA--AGCTTGGGAG---------AT--ACCGTGGGC---AGGAGCG-GGCTTTCTGATCGACG
#=GS CryWin-Amy-Flo/1-256 DE AWGH01000002.1:c940720-940467 Cryptococcus wingfieldii CBS 7118 supercont1.2, whole genome shotgun sequence; MEKH01000001.1:c1912534-1912281 Cryptococcus amylolentus CBS 6273 supercont2.1, whole genome shotgun sequence; RRZH01000002.1:c2035945-2035692 Cryptococcus floricola strain DSM 27421 chromosome 2, whole genome shotgun sequence
CryWin-Amy-Flo/1-256 ATCGACCTTA----------CTCTGCAGGATC-TCTGTATAGTAATTGTTCT--TTTTCGCTAGTCCACACTAGCCACATTTTCCACGTCGAGGGGATGATCAAGTTACCCGCCCC---GCTCTTTCACAGAGGCATTTAGCCA-CCGACA---CTTCCGGCT-----TC-----GGTCA-GTAG--C-TGTTGGGAGGTTATCGAAGACGCTGGCGGTACGT---------ATACC---GACT-GA---------TCCT--TC--GGGA----------AA--GGTCTGGGA----------ACG-GCCGTGGGC---TGGGGCG-GGCTTTCTGATCGACG
#=GS exonic-boxCDRNA/1-253 DE with-leader-251nt_upstream-snR13 ncrna 3:1423432:1423686:-1
exonic-boxCDRNA/1-253 ATCGACCTTACTC----------TGCAGGATC-TCTGTATAGTAATCGTTCT--TTTTTGCTAGTCCACACTAGCCACATTTTCCACGTCGAGGGGATGATCTAGTCACCCGCTTC---GCTCTTTCACAGAGGCATTTAGCCG-TCGG--TC--CTC-TCTT-----TC-----GAGA--GAG--GTT--CCGGGAGGTTATCGAAGAAGCTGGCGGT--AT--------GGTACC-AGTTTT-GA---------CCTC--TT--GGGG------------AAAGCTTGGGAG---------AT--ACCGTGGGC---TGGAGCG-GGCTTTCTGATCGACG
#=GS KwoMan/2-274 DE ASQF01000028.1:c358165-357892 Kwoniella mangroviensis CBS 8886 cont1.28, whole genome shotgun sequence
KwoMan/2-274 GCTGACTTTATCTGTTTGGCATATACAGGATT-TCTGTATAGTTACTGTCTCT-TTTTTGCTAGTCCATACTAGCCACATTTTCCACGTCGAGGTGATGATAAAGTTACCCATCC-T--ACTCTTTCACAGAGACATTTAGCCG-CTTCC-C-------TTTCTC--TTTC--GGGAGA-------TTA-GGAAGGAGGTTATCGAAGAAGCTGGCGAT--TC--ACGT-----GGA--CTC----A-TCTTT---ATCC-CTCGTGGAT---TTAGA--A--GAG----GG----ACG----GA--ATTGTGGGT--TT-GGATGTGGCTTTCTGATCGACG
#=GS KwoSha/10-276 DE NQVO01000013.1:c300904-300629 Kwoniella shandongensis strain CBS 12478 scaffold00013, whole genome shotgun sequence
KwoSha/10-276 GCTGACATGGATTCTCCCACTCTCACAGGATT-TCTGTATAGTCACTGTTCT--TCTTTGCTAGTCTACACTAGCCACATTTTCCACGTCGAGGTGATGATCCAGTCACCCGCCCC---GCTCTCTCACAGAGGCATTTAGCCG-TCAG----TCAAA-GTCTCTC--TC--GGGAGGTC-TTTGA-----TTGGGAGGTTATCGAAGAAGCTGGCGGTACGC-----------GGA-CCCGTC-GA---------CCTT--TC--GAGG----------AAGATGGG--GG-----------GCG-GCCGTGGGC---TGGGGCA-GGCTTTCTGATCGACG
#=GS TreMes/14-259 DE AFVY01000137.1:32273-32531 Tremella mesenterica DSM 1558 strain Fries TREMEscaffold_3_Cont137, whole genome shotgun sequence
TreMes/14-259 GCTGACCT---------------TTCAGGATT-TCTGTATAGTTATCGTCTCG-TCTTTGCTAGTCCACACTAGCGACAAATTCCACGTCGAGGTGATGAT-CACCC-CCCGCTC---CGCTCTCTCACAGAGGCCTTTGG--A-CCCGTG--TTTT-GTCATTTC-TTTT-GAGGTGA-A-GAAA---TACGGG---TCTATCGAAGAAGCTGGCGAC---------------GAA---TCT--GAC------CTTTTC-GAAA-GAAGAG-------TGA-GGA---CGA---------------GTCGCGGGCG--A-GGGCG-GGATTTCTGATCGAC-
#=GS VanFra/7-241 DE BEDY01000008.1:19565-19808 Vanrija fragicola DNA, scaffold: scaffold_8, strain: JCM 1530, whole genome shotgun sequence
VanFra/7-241 GCTTACCCTTC------------TCCAGGATC-TCTCTACAGTTTACGTCTCT-TTCTCGCTAGTTCATACTAGCCACATTTTCCACGTCGAGGCGATGATACAGTC-CCCTCCCG---GCTCTCTTACAGAGGCC--CG---AG-CCCA-CT-------GCTT----TC--GAGC---TT----------TGGG-A---CG-CGAAGAAGCTGGCGG-C-GC--TGG----ACAAA---CCC--GAC--------CCCG--CG--TGGG----------AA-GGG---CGAGA---CC----GC-A-CCGTGGGC---ACGGGAG-GGCTTTCTGATCGACG
#=GS VanHum/1-233 DE BCJF01000006.1:c522088-521856 Vanrija humicola DNA, scaffold: scaffold_5, strain: JCM 1457, whole genome shotgun sequence
VanHum/1-233 -------CATGTGTCTATACTC-AGTAGGATC-TCTCTACAGTATACGTCTCA-TTCTCGCTAGTTCATACTAGCCACATTTTCCACGTCGAGGCGATGATCTAGCC-CCCTCCCG---GCTCTCTTACAGAGGACCC------CGAG------------CGTCT---TC---GGACG--T-----------CTC-A-----TCGAAGAAGCTGGCGGC--G---AGG----ACGA---GCCC--GA----------CCC-CCTGTGGG-----------AA-GGGC---GAGG---CCG----C--GCCGTGGGC---TCGGGAG-GGCTTTCTGATCGACG
#=GS VanHum/35-266 DE QKWK01000005.1:c902259-901991 Vanrija humicola strain CBS 4282 CBS4282_scaffold05, whole genome shotgun sequence
VanHum/35-266 GTTTCCCCATGTGTCTATACTC-AGTAGGATC-TCTCTACAGTATACGTCTCA-TTCTCGCTAGTCTACACTAGCCACATTTTCCACGTCGAGGCGATGATCTAGC--CCCTCCCG---GCTCTT-CACAGAGGACCC---------------------GGGCGC--CTA---GCGCCC-----------------A-----TCGAAGAAGCTGGCGGC-GGGT----------GA---GTCT--GA--------CCCCC-GCAA-GGGGG---------GA-GGGC---GA----------GCCT-GCCGCGGGC---TTGGGAG-GGCTTTCTGATCGACT
#=GS ApiSia/17-245 DE JALJEG010000053.1:46680-46928 Apiotrichum siamense strain L8in5 NODE_53_length_158554_cov_10.826822, whole genome shotgun sequence
ApiSia/17-245 GCTGACCCGCC------------TACAGGATC-TCTCTACAGTAAACGTCTCT-TTCTCGCTAGTTCATACTAGCCACATTTTCCACGTCGAGGCGATGATACAGCC-CCTTCCCG---GCTCTTTCACAGAGCGCT-C--------------ACCC--GCCGT----CC----GCGGC--GGGT-----------A----G-CGAAGAAGCTGGCGGC--G---CGCGCC---GA---GCCC--GA----------CCT--CT--AGG---------GGAA-GGGC---GA--GGCGC-----C--GCCGTGGGC---ACGGGAA-GGCTTTCTGATCGACT
#=GS ApiAki/24-253 DE PQXP01000021.1:c81431-81179 Apiotrichum akiyoshidainum strain HP2023 Contig740, whole genome shotgun sequence
ApiAki/24-253 GCTCACACCT-------------CCCAGGATC-TCTCTACAGTAAACGTCTCT-TTCTCGCTAGTTCATACTAGCCACATTTTCCACGTCGAGGTGATGATACAGTC-CCCTCCC---TGCTCTTTCACAGAGCGCT-C------GC----A---AC---TGTGC---TCT--GCACG---GT----A-----GC-A----G-CGAAGAAGCTGGCGGC-GGAC---------ACAAA--CTC--GA----------CCT--CC--AGG---------GGAA-GAG---CGCGC--------GTCT-GCCGTGGGCA--C-GGGAG-GGCTTTCTGATCGACT
#=GS CutCur/10-231 DE NIUX01000029.1:77100-77334 Cutaneotrichosporon curvatum strain ATCC 10567 Contig029, whole genome shotgun sequence
CutCur/10-231 GCTGACCCA--------------CGCAGGATC-TCTCTACAGTCGCTGTTCAA--TTTCGCTAGTCTACACTAGCCACATTTTCCACGTCGAGGTGATGATCGAGC--CCCTCGC-A--GCTCTTTCACAGAGCGCT-TGCCC---CGG-----------TC------TC------GG-------------CTG-T-GGGCAGCGAAGAAGCTGGCGGCG-----AGTGC----AA----CCC-CGA----------CC--TCTT--GG-----------GA-GGG---CGC---GCGCA------AGCCGTGGGC---C-GCGAG-GGCTTTCTGATCGACT
#=GS TriOvo/30-261 DE JAMFRF010000027.1:530619-530898 Trichosporon ovoides strain 2NM914A ctg_27, whole genome shotgun sequence
TriOvo/30-261 GCTAACGTTC-------------TCCAGGATTCTCTGTATAGTCACTGTCTT--TTCTCGCTAGTTCACACTAGCCACATTTTCCACGTCGAGGAGATGATCTAGTC-CCCTCCCG---GCTCTTTCACAGAGACCAC------T------TT--GCA-ACACC----TCCG--GGTGTC-TGC-----------T------CCGAAGAAGCTGGCGGC--GC--TGGC----AGAA---CCT--GAC---------CC--TCTGT-GG-----------AA-AGG---CGAC---GCC----GC--GCCGTGGGC---ACGGGAG-GGCTTTCTGATCGACG
#=GS KocImp/58-286 DE NBSH01000010.1:c93067-92780 Kockovaella imperatae strain NRRL Y-17943 BD324scaffold_10, whole genome shotgun sequence
KocImp/58-286 GCTGACTCGTCT-----------CTCAGGATT-TCTGTATAGTTAGTATTTA--TCTTTGCTAGTCTACACTAGCCACATTTTCCACATCGAGGGGATGATCCAGTA-CCCGCTC---CGCTCTT-CACAGAGACATCTTGAC--GTG-------------------CTCCT--------------------CGC-CGTCAATCGAAGAAGCTGGCAGG--GA---------------TCGTC--AA------TCGACTC-TTCACGGGTTGA-------GC-GACGG-T-------------TT--CCTGTGGGCG-AT-GAGCG-GGCTTTCTGATCGATG
#=GS DioHun/20-247 DE JAMRJJ010000003.1:2151490-2151740 Dioszegia hungarica strain Y1 Contig3, whole genome shotgun sequence
DioHun/20-247 GCTGACCCC--------------CTCAGGATT-TCTGTATAGTCACTGTTCT--CTTATGCTAGTCCACACTAGTCATAAATTCCACGTCGAGGCGATGATACAGTA-CCCGCTC-G--GCTCTCTTACAGAGGCATTTCC--AG------A-TCGC--TC-------TCG---G---G-T-GTGA----------C--GGATCGAGGACGCTGGCGGT-TGA--TGTT-----GA---GCT---GA---------GCTC--TA--GGGC----------AA-GGC----AA----GAC----TCG-GCTGTGGGC---A-GAGCG-GGCTTTCTGATCGACG
#=GS CryFla/20-248 DE CAUG01000664.1:c55436-55185 Cryptococcus flavescens NRRL Y-50378 WGS project CAUG00000000 data, contig NODE_2187_length_71177_cov_45_840343, whole genome shotgun sequence
CryFla/20-248 GCTGACCC---------------CGCAGGATT-TCTGTATAGTAAATGTCTCG-TTTTTGCTAGTCTACACTAGCCACATTTTCCACGTCGAGGGGATGATTTAGTA-CCCGCTC---CGCTCTTTCACAGAGACATTTGG--C-CTC-----------GCCC-----TC-----GGGC--------GG---GAG-A--CCATCGAAGAAGCTGGCGAC-A-------------GGA--GC---CGAT------CTACCC-CTTGTGGGTGG------TGAC-GC----CGA-------------T-GTCGCGGGCG--A-GAGCG-GGCTTTCTGATCGACG
#=GS TriGra/6-227 DE BCJO01000001.1:4619041-4619267 Trichosporon gracile DNA, scaffold: scaffold_0, strain: JCM 10018, whole genome shotgun sequence
TriGra/6-227 GCTCACAC-C-------------ACCAGGATC-TCTCTACAGTAAACGTCTCT-TTCTCGCTAGTTCACACTAGCCACATTTTCCACGCCGAGGTGATGATCCAGT--CCCCCCAG---GCTCTTT-ACAGAGCGCG---------CGG----------CGCGTC--CTG---GACGCG------------CCG-------G-CGAAGACGCTGGCGT-AGCGC----------GAG-CGCTG--GA----------CCT--TCT-GGG----------GAA-CGGCG-CGA----------GCGTG-GCGTGGGC---TCTGGGG-GGCTTTCTGATCGGC-
#=GS TriLai/5-234 DE BCKV01000009.1:1266798-1267031 Trichosporon laibachii DNA, scaffold: scaffold_8, strain: JCM 2947, whole genome shotgun sequence
TriLai/5-234 GCTGACCCCCT------------ACCAGGATC-TCTCTACAGTTAACGTCTCT-TTCTCGCTAGTTCATACTAGCCACATTTTCCACGTCGAGGTGATGATACAGTC-CCCTCCC---TGCTCTTTCACAGAGCGCTA-------GC----G---AC-A--GTGC---TCT--GCAC--A-GT----A-----GC-A-----GCGAAGAAGCTGGCGGCGGACGC---------AA---ACTC--GA----------CCT--CC--AGG---------GGAA-GAGT---GC---------GCGTCGGCCGTGGGCA--A-GGGAG-GGCTTTCTGATCGAC-
#=GS TriLai2/6-235 DE BCKV01000009.1:395072-395306 Trichosporon laibachii DNA, scaffold: scaffold_8, strain: JCM 2947, whole genome shotgun sequence
TriLai2/6-235 GCTGACCCCCT------------ACCAGGATC-TCTCTACAGTAAACGTCTCT-TTCTCGCTAGTTCATACTAGCCACATTTTCCACGTCGAGGTGATGATACAGTC-CCCTCCC---TGCTCTTTCACAGAGCGCT-C------GCG-------AC---TGTGC---TCT--GCACA---GT----A-----GC-A----G-CGAAGAAGCTGGCGGCGGACGC---------AAA---CTC--GA----------CCT--CC--AGG---------GGAA-GAG---CGC---------GCGTCGGCCGTGGGCA--C-GGGAG-GGCTTTCTGATCGAC-
#=GS ApiDom/9-231 DE BCFW01000007.1:c390824-390594 Apiotrichum domesticum DNA, scaffold: scaffold_6, strain: JCM 9580, whole genome shotgun sequence
ApiDom/9-231 GCTCACAC---------------CTCAGGATC-TCTCTACAGTCAACGTCTCT-TTCTCGCTAGTTCATACTAGCCACATTTTCCACGTCGAGGTGATGATACAGTC-CCCTCCCG---GCTCTTTCACAGAGCGC--TGACC--CCC------------------TCTTTT--------------------GGGT-GGTCG-CGAAGAAGCTGGCGGC-GCGG----------ACGC-GCTC--GA----------CCC--CCC-GGG----------GAA-GAGC-AAGG----------CTGC-GCCGTGGGC---TCGGGAG-GGCTTTCTGATCGAC-
#=GS TriFae/6-238 DE JXYK01000003.1:1200015-1200252 Trichosporon faecale strain JCM 2941 scaffold_0003, whole genome shotgun sequence
TriFae/6-238 GCTAACTTTC-------------TCCAGGATTCTCTGTATAGTCACTGTCTT--TTCTCGCTAGTTCACACTAGCCACATTTTCCACGTCGAGGAGATGATCTAGTC-CCCTCCCG---GCTCTTTCACAGAGACCA-C-----T------TT--GC--GGCGCT--CTCGC-AGCGCT-CGC------------T------CCGAAGAAGCTGGCGGC--GC--TGGCA----GAA---CCT--GA----------CC-CTCTGT-GG-----------AA-AGG---CGA---TGCC-----GC-GCCGTGGGC---ACGGGAG-GGCTTTCTGATCGACG
#=GS SaiPod/2-238 DE RSCD01000015.1:c529658-529421 Saitozyma podzolica strain DSM 27192 scaffold_15, whole genome shotgun sequence
SaiPod/2-238 GCTGACATGCAT-----------CGTAGGATT-TCTGTATAGTAAACGTCCT--TTTTCGCTAGTCTACACTAGCCACATTTTCCACGTCGAGTTGATGATCCAGTC-CCCGCCCG---GCTCTTTCACAGAGGCATTTGGCGA-CCCG----------TCTC-----TC-----GGG-A-----------CGGG-ACGCCATCGAGGAAGCTGGCGG-TGTCG-TGCC-----GA---GCTC--GA----------CCT-TT---GGG-----------AA-GAGC---GA----GGT---CGAC--CCGCGGGC---TCGGGCG-GGCTCTCTGATCGACG
#=GS BulAlb/4-241 DE CAMYTR010000032.1:c72770-72530 Bullera alba genome assembly, contig: jcf7180000012125, whole genome shotgun sequence
BulAlb/4-241 ACTGACTCTTCT-----------CTAGGGATT-TCTGTATAGTTAATGTCTCG-TCTTTGCTAGTCTACACTAGCCACATTTTCCACGTCGAGGTGATGATCTAGTC-CCCACCC---CGCTCTTTCACAGAGACTTTTGGT-----------------TCGTTG--CTT---CGGCGA-------CGC------T--ACCATCGAAGAAGCTGGCGG-CGGTA--CGA-----------GTC--GAA-------TCGGC-CTTGTGTTGA--------CAA-GAC----------TTG---TACC--CCGTGGGCG-TT-GGGTG-GGCTTTCTGATCGACG
#=GS PapTer/5-233 DE JAHXHD010000715.1:c3078-2845 Papiliotrema terrestris strain LS28 scaffold-714, whole genome shotgun sequence
PapTer/5-233 GCTGACTC---------------TGCAGGATT-TCTGTATAGTTAATGTCTCG-TTTTTGCTAGTCTACACTAGCCACATTTTCCACGTCGAGGGGATGATTTAGTA-CCCGCTC---CGCTCTTTCACAGAGACCTTTGG-----C---------C--TCGCCC---TC---GGGCGG--G------T----G--A--CCATCGAAGAAGCTGGCGAC--A------------GGA----GC-CGAT------CTACCC-CTTGTGGGTGG------TGAA-GC----CGA------------T--GTCGCGGGCG--A-GAGCG-GGCTTTCTGATCGACG
#=GS PapLau/6-240 DE JDSR01001049.1:c26155-25916 Papiliotrema laurentii RY1 contig_1086, whole genome shotgun sequence
PapLau/6-240 CTTGACCATATTC----------TGCAGGATC-TCTGTATAGTTATCGTCTCG-TTTTTGCTAGTCTACACTAGCCACATTTTCCACGTCGAGGGGATGATCTAGTT-CCCGCCC---CGCTCTTTTACAGAGGACTTTGG-----CG-----------TTTTCT---TC---GGAAGA-------------CG--A--CCATCGAAGAAGCTGGCGG-CTGTT--TGTC---------------GA------GCTGCTCCTTTGCGGGCGGT-------AA-------------GACG---AACA--CCGTGGGCG--C-GGGCG-GGCTTTCTGATCGACG
#=GS NaeAur/2-231 DE JAKFAO010000007.1:c1617317-1617087 Naematelia aurantialba strain NX-20 Contig7, whole genome shotgun sequence
NaeAur/2-231 GCTGACCCGCC------------CGCAGGATT-TCTGTATAGTTATTGTCTCG-CTTTTGCTAGTCTACACTAGCCATACTATCCACGTCGAGGTGATGATCGAGTC-CCCGCC---CCGCTCTTTCACAGAGACCTCTGGT--CGTCC------TTC----------TC-----------GAG-------GGAC-A-GCCATCGAAGAAGCTGGCGGT-GTC---GGCT--------------GGAT------CTCTTC--TC--GGAGAG------TGAA-------------GGCT----GGC--CCGTGGGCGG-C--GGCG-GGCTTTCTGATCGACG
#=GS NaeEnc/4-240 DE MCFC01000048.1:c73912-73673 Naematelia encephala strain 68-887.2 BCR39scaffold_48, whole genome shotgun sequence
NaeEnc/4-240 AGTGATTATGCTT----------CGCAGGATT-TCTGTATAGTATATATCTCA-CTTTTGCTAGTCTACACTAGCCACACTATCCACGTCGAGGTGATGATCGAGTC-CCCGCC---CCGCTCTTTCACAGAGACTTTTGGT--CGTTC------CTC--------CTTGTT---------GGG-------GAAC-A-ACCATCGAAGAAGCTGGCGG--TTG-----------TTG---ATT-GGAT------CTCTCT--TC--GGGGAG------TGAA-GGT-----------------CAAC-CCGTGGGCGG-C--GGCG-GGCTTTCTGATCGACG
#=GC SS_cons -----------................................................((((((....))))))-----------[[[[[[---<<<<---------[[[[[[[[.[[[[[[-----<<<<------(((((..((((((..(((((.(((((((......)))))))..)))))...))))))..)))))-->>>>------(((((.(((((.(((((.......((((((....(((..(((((((......)))))))..)))....))))))......)))))..))))).)))))]]]]]]..]]]]]]-]]....>>>>]]]]]]
//
Image source U13:
# STOCKHOLM 1.0
#=GF RW van Nues, May 2023
#=GF https://coalispr.codeberg.page/paper
#=GS hU13/1-104 DE gi|94721317|ref|NR_003041.1| Homo sapiens small nucleolar RNA, C/D box 13 (SNORD13), small nucleolar RNA
hU13/1-104 ---ATCCTTTTG--TAGTT------CATGA-GCGTGATGATTGGGTG---------------T-TCATAC-----GCTT------GTGTGAG--ATGTGCC-----ACCCTTG--AACCTTGTTACGACGTG------GGCACAT--------------------TACC-CG-TCTGACC-----
#=GS MusMus/1-108 DE from gi|12841307|dbj|AK007639.1| Mus musculus 10 day old male pancreas cDNA, RIKEN full-length enriched library, clone:1810029E06 product:unclassifiable, full insert sequence
MusMus/1-108 -GGATCCTTTC---TGGTT------CATAAAGCGTGATGATTGGGTG---------------T-TCACGC-----CATT------GCGTGAC--ATGTGCC-----GCCCATA--AACCTTGTTACGACGTG------GGCACAT--------------------TACC-CG-TCTGACATG---
#=GS MusMus/1-109 DE from gb|AC157667.11|:162770-162890 Mus musculus 6 BAC RP24-86B22 (Roswell Park Cancer Institute (C57BL/6J Male) Mouse BAC Library) complete sequence
MusMus/1-109 -GGATCCTTTC---TGGTT------CATAAAGCATGATGATTGGGTG---------------T-TCACGC-----CGTT------GCGTGAC--ATGTGCC-----GCCCATA--AACCTTGTCATGGCGTG------GGCACAT--------------------TACC-CG-TCTGACGTGA--
#=GS FelCat/1-109 DE from gi|586983258:c347-180 PREDICTED: Felis catus TELO2 interacting protein 2 (TTI2), transcript variant X2, mRNA
FelCat/1-109 --GATCCTTTTG--TAGTT------CATAA-GCGTGATGATTGGGTG---------------T-TCACGC-----CGTT------GTGTGAC--ATGTGCC-----ACCCTTA--AACCTTGTTACGACGCT------GGCACAT--------------------TACC-CG-TCTGACGTGAA-
#=GS AciJUba/1-109 DE from gi|961715954:c186-74 PREDICTED: Acinonyx jubatus TELO2 interacting protein 2 (TTI2), transcript variant X3, mRNA
AciJUba/1-109 --GATCCTTTTG--TGGTT------CATAA-GCGTGATGATTGGGTG---------------T-TCACGC-----TGTT------GTGTGAC--ATGTGCT-----ACCCTTA--AACCTTGTTACGACGCT------GGCACAT--------------------TACC-CG-TCTGACGTGAA-
#=GS UrsMar/1-109 DE from gi|671036750:68-180 PREDICTED: Ursus maritimus uncharacterized LOC103681354 (LOC103681354), ncRNA
UrsMar/1-109 --AAAAATATCG--AAGTT------CATGA-GCGTGATGATTGGGTG---------------T-TCACGC-----TGTT------GTGTGAC--ATGTGCC-----TCCCTCA--AACCTTGTTACGACCTG------GGCACAT--------------------TACC-CG-TCTGACGTGAA-
#=GS CanLup/1-108 DE from gi|928156847:c220-100 PREDICTED: Canis lupus familiaris TELO2 interacting protein 2 (TTI2), transcript variant X2, mRNA
CanLup/1-108 --GATCCTTTTG--TAGTT------CATAA-GCGTGATGATTGGGTG---------------T-TCACGC-----CGTT------GTGTGAC--ATGTGCC-----TCCCTCC--AACCTTGTTACGACG-C------GGCACAT--------------------TACC-CG-TCTGACGTGAA-
#=GS SusScr/1-109 DE from gi|927209768:c1317-1172 PREDICTED: Sus scrofa TELO2 interacting protein 2 (TTI2), transcript variant X2, misc_RNA
SusScr/1-109 --GATCCTTTTG--TAGTT------CATAA-GCATGATGATTGGGTG---------------T-TCACGC-----GCCT------GCGTGAT--ATGTGCC-----TCCCTCA--AACCTTGTTACGACATC------GGCACAT--------------------TACC-CG-TCTGACGTGAA-
#=GS HomSap/1-111 DE from gi|508772642:c4720-4575 Homo sapiens TELO2 interacting protein 2 (TTI2), RefSeqGene on chromosome 8
HomSap/1-111 ATGATCCTTTTG--TAGTT------CATGA-GCGTGATGATTGGGTG---------------T-TCATAC-----GCTT------GTGTGAG--ATGTGCC-----ACCCTTG--AACCTTGTTACGACGTG------GGCACAT--------------------TACC-CG-TCTGACCTGAA-
#=GS NomLeu/1-109 DE from gi|820995404:c134-1 PREDICTED: Nomascus leucogenys TELO2 interacting protein 2 (TTI2), transcript variant X1, mRNA
NomLeu/1-109 --GATCCTTTTG--TAGTT------CATGA-GTGTGATGATTGGGTG---------------T-TCATAC-----GATT------GTGTGAG--ATGTGCC-----ACCCTTA--AACCTTGTTACGACGCG------GGCACAT--------------------TACC-C-TTCTGACGTGAA-
#=GS CapHir/1-111 DE from gi|926725765:c588-450 PREDICTED: Capra hircus TELO2 interacting protein 2 (TTI2), transcript variant X3, mRNA
CapHir/1-111 --GATCCTTTTG--TAGTT------CATGA-GCGTGATGATTGGGTG---------------T-TCACGC-----TGCT------GCGTGAC--ATGTGCC-----TCCCCGCTAAACCTTGTTACGACACC------GGCACAT--------------------TACC-CG-TCTGACGTGAA-
#=GS GorGor/1-111 DE from gi|426359305:c614-469 PREDICTED: Gorilla gorilla gorilla TELO2 interacting protein 2, transcript variant 1 (TTI2), mRNA
GorGor/1-111 ATGATCCTTTTG--TAGTT------CATGA-GCGTGATGATTGGGTG---------------T-TCATAC-----GCTT------GTGTGAG--ATGTGCT-----ACCCTTG--AACCTTGTTACGACGTG------GGCACAT--------------------TACC-CG-TCTGACCTGAA-
#=GS MacMul-SNORD13/1-108 DE gi|284807305|gb|FJ915995.1| Macaca mulatta C/D box 13 snoRNA, complete sequence
MacMul-SNORD13/1-108 --GATCCTTTTG--TGGTT------CATGA-GCATGATGATTGGGTG---------------T-TCACAC-----GTTT------GTGTGAG--ATGTGCC-----ACCCTAC--AACCTTGTTACGACGTC------GGCACAT--------------------TCCC-CA-TCTGACCTGA--
#=GS PonPyg/1-109 DE from gi|1043015352:c64348309-64348180 Pongo pygmaeus genome assembly, chromosome: XII
PonPyg/1-109 ---ATCCTTTTG--AAGTT------CATAA-GCATGGTGATTGGGTT---------------T-TCACAC-----TCAT------GCGTGAG--ATGTACC-----ACCCTCA--AAGCTTGTTATGATGTG------GGCACAT--------------------TACC-CA-TCTGACATGAGA
#=GS SNOR105/1-129 DE gi|240254421:10151668-10151774 Arabidopsis thaliana chromosome 1
SNOR105/1-129 ATGATCCTTCAGGCAAGTT--AAAGGGG---ATATGATGAATG-GTA----------AAAA----CTCGC---TTATATT-----GCGAG-AA-GAGCGTT-CCG--CCCAA---AGCCT-GTTACGACTATT--GG-GCGCTCTCT----TTTT----------TAC-ACAATCTGAT-CCCTC
#=GS SNOR108/1-130 DE gi|240256493:16598796-16598905 Arabidopsis thaliana chromosome 5, complete sequence
SNOR108/1-130 ATGATCCTTCAGGCAAGTT--ATAGGGG---AAATGAGGAATG-GT------TAT---AAT----CTCGC---TTTAAT------GCGTG-AT-GAGCGCT-CCG--CTCAA---AGCCT-GTTACGACTAATT-GG-GCGCTC-CA----ATT--TTT-------AT-ACAATCTGAT-CCCTC
SNOR146/1-127 ATGATCCTTCAGGCAAGTT--AAAGGGG---AAATGAGGAATG-GT------TATTA-AA-----CTCGC----TTAAT------GCGAG-A-AGAGTGTT-TCG--CTC-----AGCCT-GTTACGACTATT--GA-GCACTCTC-C----TT-CTTTT------AT--CA-TCTGAT-CCCTC
#=GS SolPen_/1-132 DE gi|663673442:76110346-76110477 Solanum pennellii chromosome ch04, complete genome
SolPen_/1-132 ATGATCCTTCAGGCAAGTTTTGAT-GGG---ATATGATGAGTG-GT-------C----AA--TA-CTCGCA--ACAAAATA---TGCGAG-AT-GGGCGTT-CCAG-CTC-----AGCCTGGTTACGACAATT-TGG-GCGCTCTCG-----TT-CAA--------AC--CAATCTGAT-CCCT-
#=GS CamSat/1-130 DE gi|727485057:c210-81 PREDICTED: Camelina sativa methyltransferase-like protein 23 (LOC104704008), mRNA
CamSat/1-130 ATGATCCTTCAGGCAAGTT--AAAGGGG---AAATGAGGAGTG-GT-TAAAA------AA--T--CTCGC----TCTAAT-----GCGAG-A-AGAGTGTA-TCG--CTC-----AGCCT-GTTACGACAATTT-GA-GCACTCTC-C----TT-C----TTTT--GT--CA-TCTGAT-CCCTC
#=GS CamSat2/1-127 DE gi|727601320:c158-32 PREDICTED: Camelina sativa methyltransferase-like protein 23 (LOC104753054), mRNA
CamSat2/1-127 ATGATCCTTCAGGCAAGTT--AAAGGGG---AAACGAGGAGTG-GT--------TT--AA--T--CTCGC----TTTATT-----GCGAG-A-AGAGTGTA-TCA--CTC-----AGCCT-GTTACGACAATTT-GA-GCACTCTC-C----TT-CTTTT------GT--CA-TCTGAT-CCCTC
#=GS BraRap/1-131 DE gi|685355907:c136-6 PREDICTED: Brassica rapa methyltransferase-like protein 23 (LOC103838247), mRNA
BraRap/1-131 ATGATCCTTCAGGCAAGTG--AAAGGGG---AAATGAGGAGTG-GT-T----------AT-TTT-CTCGC---TTTTATTAT---GCGAG-A-AGAGCGTT-TCG--CTC-----CGCCT-GTTACGACAATT--GA-GCGCTCTC-CTTT-GT-CTT--------AC--CA-TCTGAT-CCCTC
#=GS BraNap/1-131 DE gi|923799832:c137-7 PREDICTED: Brassica napus methyltransferase-like protein 23 (LOC106391010), transcript variant X2, mRNA
BraNap/1-131 ATGATCCTTCAGGCAAGTG--AAAGGGG---AAATGAGGAGTG-GT-T-----ATT--TA-----CTCGC---TTTTATTAT---GCGAG-A-AGAGCTCT-TCG--CTC-----CGCCT-GTTACGACAATT--GA-GCGCTCTC-CTT--TG-TCTT-------AC--CA-TCTGAT-CCCTC
#=GS SolPen/1-131 DE gi|663673442:76072650-76072780 Solanum pennellii chromosome ch04, complete genome
SolPen/1-131 ATGATTCTTCAGGCAAGTTTTAAC-GGG---ATGTGATGAGTG-GT-------C----AAT--A-CTCGCAT--TAAAAT---ATGCGAG-A-AGAGTGTC-CAGG-CTC-----AGCCT-GTTACGACAATT-TGG-GCACTCTC-A---ATT---TA-------AC--CAATCTGAT-CCCT-
#=GS SolPen3/1-131 DE gi|663673439:89633578-89633708 Solanum pennellii chromosome ch01, complete genome
SolPen3/1-131 ATGATCCTTCAGGCAAGTTTTAAT-GGG---ATGTGATGAGTG-GT-----------CAAT--A-CTCGCA--ATAAAAA----TGCGAG-A-AGAGCGTT-CCA--GCTC----AGCCTGGTTACGACAATTT-GG-GCGCTCTC-----ATTG--TT-------AC--CAATCTGAT-CCCT-
#=GS SolPen4/1-131 DE gi|663673439:89635752-89635882 Solanum pennellii chromosome ch01, complete genome
SolPen4/1-131 ATGATCCTTCAGGCATGTTTTAAT-GGG---ATGTGATGAGTG-GT-------CAT--AA-----CTCGCA--ATAAAAA----TGCGAG-A-AGAGCGTT-CCA--GCCC----AGCCTGGTTACGACAATTT-GG-GCGCTCTC-A----TT---TTT------AC--CAATCTGAT-CCTT-
#=GS SolLyc3/1-131 DE gi|663680609:71983035-71983165 Solanum lycopersicum chromosome ch01, complete genome
SolLyc3/1-131 ATGATCCTTCAGGCAAGTTTAAAT-GGG---ATGTGATGAGTG-GT-------CAT--TA-----CTCGCA--ATAAATA----TGCGAG-A-AGAGCGTT-CCA--GCCC----AGCCTGGTTACGACAAAAT-GG-GCGCTCTT-AT---TA---TT-------AC--CAATCTGAT-CCCT-
#=GS SolLyc4/1-130 DE gi|663680609:71985354-71985483 Solanum lycopersicum chromosome ch01, complete genome
SolLyc4/1-130 ATGATCCTTCAGGCAAGTTTTAAT--GG---ATGTGATGAGTG-GT-------CAT--AA-----CTCGCA--ATAAAAA----TGCGAG-A-AGAGCGTT-CCA--GCCC----AGCCTGGTTACGACAATTT-GG-GCGCTCTT-AA---TT---TT-------AC--CAATCTGAT-CCTT-
#=GS SolLyc1/1-132 DE gi|663680879:63071585-63071716 Solanum lycopersicum chromosome ch04, complete genome
SolLyc1/1-132 ATGATCCTTCAGGCAAGTTTTAAT-GGG---ATATGATAGGTG-GT-------C----AA--TA-CTCGCA--ATAAAATA---TGCGAG-AT-GGGCGTT-CCA--GCTC----AGCCTGGTTACGACAATTT-GA-GCGCTCTCG-----TT-CAA--------AC--CAATCTGAT-CCCT-
#=GS OryJap/1-131 DE gi|937937345:c5895114-5894984 Oryza sativa Japonica Group DNA, chromosome 11, cultivar: Nipponbare, complete sequence
OryJap/1-131 ATGATCCTTCAGGCA-GTT--AAA-GGG---AGATGATGAGTG-GT--AA---C-----AT----CTCGC--TAATTAATGTT--GCGAG-A-GGGGTGT---CTGCCCCAACCAAGCCT-GTTACGACAA---CAGGGCACTCC--A---AT--ATT----TT--AC--CA-TCTGAT-CCC--
#=GS OryCoa/1-260 DE gb|GQ203301.1|:32697-32826 Oryza coarctata clone OC_Ba202M20, complete sequence
OryCoa/1-260 ATGATCCTTCAGGCA-GTT--AAA-GGG---AGATGATGAGTG-GT--AA---CA----A-----CTCGT--TAATTTTGTT---GCGAG-A-GGGGTGT---CTGCCCCAACTAAGCCT-GTTACGACAA---CAGGGCACTCC--A----T--CCAT---TT--AC--CA-TCTGAT-CCC--
#=GS MedTru/1-124 DE gb|AC137065.26|:13579-13702 Medicago truncatula clone mth2-29h7, complete sequence
MedTru/1-124 ATGATCCTTCAGGCA-G----AACGGGG---AAATGAGGAGTG-G--T---------TGG--AT-CTCAC---CAATTTT-----GTGAG-T-TGAGTGT-TCC---GCCCAC--AGCCTGGTTACGACAATT--GG-GCACTC---TC---CCA--ATT-------C--CAATCTGAT-CCCT-
#=GC SS_cons ........................<<<<...<...[[[[[..(.(((.((((.....((((...((((((((...........)))))))).(((((((((.((((............................))))))))))).)....))))......)))).))).)).]]]]]>.>>>>.
#=GF R2R keep allpairs
//