U3 or snR45¶
- In yeast S. cerevisiae the abundant snoRNA U3, which is essential for ribosome biogenesis, is expressed by two genes (snR17a and snR17b). Two ncRNA molecules can be identified in Cryptococcus that appear to be U3 homologues based on the RFAM (Rfam RF00012) annotation for one (URS0000D98D81_71784) of their Tremellomycetes’ counterparts. The sequences of both U3-like molecules, snR45-U13/U3a and U3b (expressed from genes on chromosomes 3 and 8 in C. deneoformans or 3 and 14 in C. neoformans), are nearly identical:
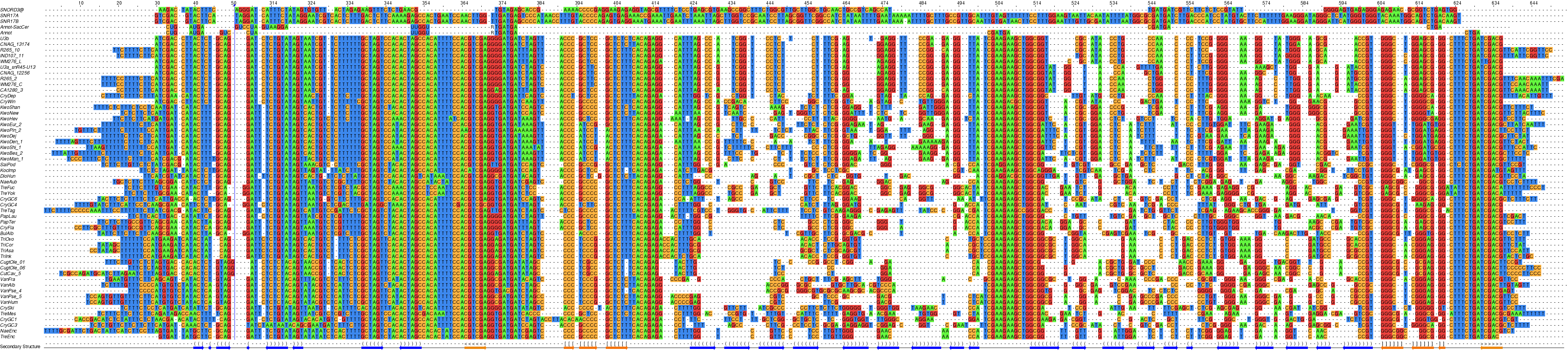
U3 homologues in Tremellomycetes, nucleotide coloring.¶
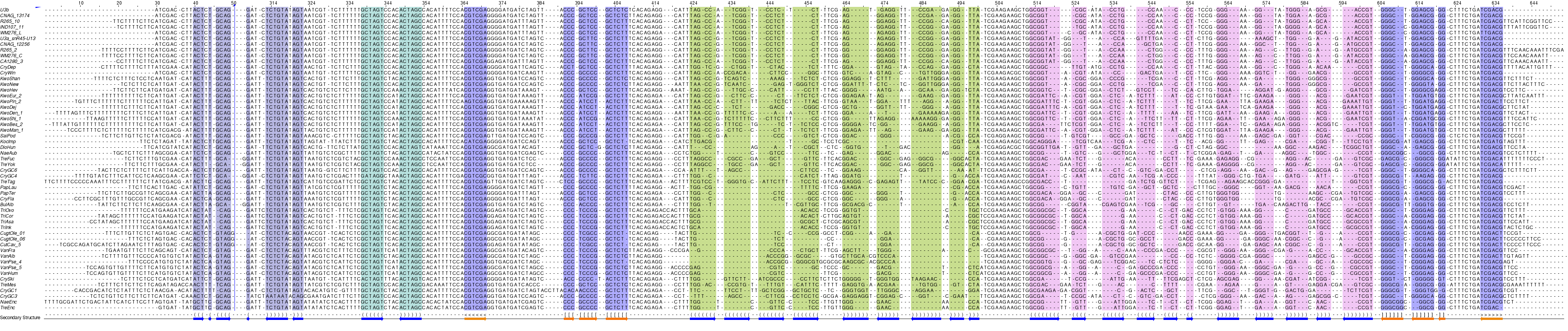
snR45/U3 homologues in Tremellomycetes, helix coloring.¶
Despite their homology to yeast U3 snoRNAs, the Cryptococcus molecules deviate with respect to their relatively short 5’ stem-loop, while in yeast U3 (RF01846) this is a major structural element comprising box A' and box A ([Samarsky-1998], see their U3 model below; in which Box C'/D is called C/D for other snoRNAs and Box B/C, D’/C’; 5'ETS is ETS1 and 3'ETS, ETS2).
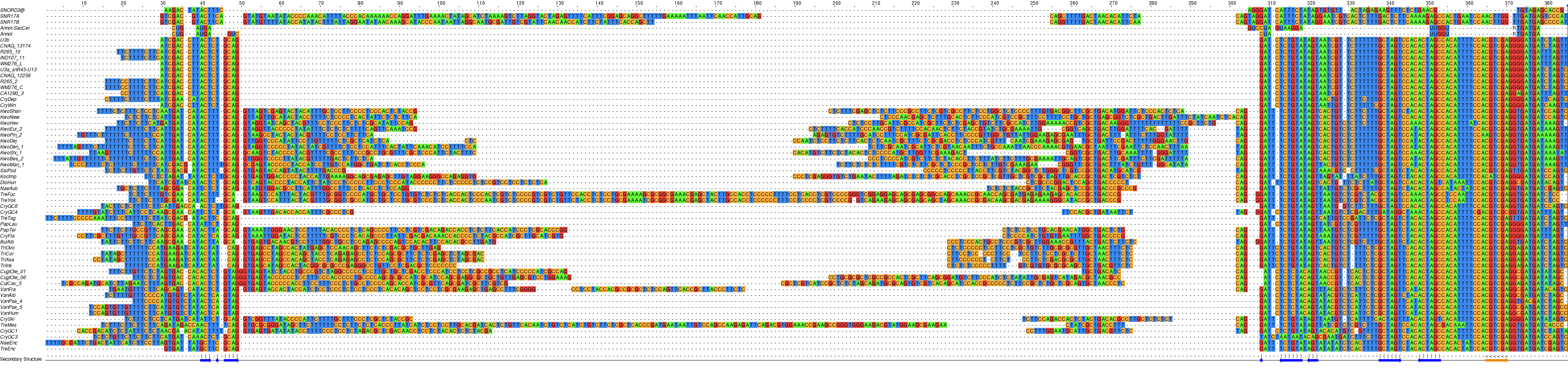
Like in S. cerevisiae, some snR45/U3 genes in Tremellomycetes are split by an intron, separating the 5’ end with box A' from the remainder of the snoRNA (only the 5’ region of U3 is shown).¶
The box A' and box A elements in yeast U3 confer complimentarity to 18S rRNA sequences forming an essential pseudo-knot in the mature rRNA. The U3 snoRNAs of Cryptococcus target through their 72 nt leader identical 18S rRNA sequences, tgatcctg[cc]agtagtc (Helix II and Helix III) as in S. cerevisiae (18S target 1a and target 1b) (See model and for comparison [Marmier-Gourrier-2011]). In baker’s yeast another 18S rRNA target, aggaa (Helix I), and interaction sites in the ETS1, Tcaaagagtg (Helix V) and ggatttggtgg (Helix VI), have been described. ETS1 sequences that could support comparable basepairing with U3 in Cryptococcus can be found: gtgaggaagaa (Helix V) and gtggaaggtg (Helix VI).
In Tremellomycetes the section of the box A U3-element that would interact with 18S rRNA pseudo-knot sequence tcct in target tgatcctg is present in the helical structure while box A' seems partly incorporated, resulting in a shorter 5’ stemloop. For Cryptococcus, however, a longer 5’ stemloop accommodating box A' can be modeled. Still, the sequence changes might affect the proposed interaction with the pseudo-knot sequence aggaa in Helix I, possibly altering the role of U3 in the assembly of the 18S rRNA pseudo-knot in Cryptococcus.
Putative interactions between U3 snoRNA and pre-rRNA in C. deneoformans as based on [Marmier-Gourrier-2011]. The box A'- and box A-like elements differ from those in yeast; Helix I is less well supported (by three instead of four basepairs) because of nucleotide changes in between the two conserved sections of box A.¶
Maybe, in Tremellomycetes, the 5’ leader sequence has other functions. In baker’s yeast, not only U3, but also snoRNAs snR4 and snR45 (U13 in higher eukaryotes) contain a leader sequence with base-pairing functionality to 18S rRNA. These snoRNAs associate with Kre33/NAT10 (CNN01050, CNAG_06379) and guide this acetyltransferase to target cytosines that get modified during maturation of the small ribosomal subunit [Sharma-2017], supposedly through a mechanism similar to pseudouridylation by H/ACA snoRNAs.
In contrast to Kre33, which is essential for ribosome biogenesis, no obvious homologues of snR45/U13 or snR4 could be identified in Cryptococcus by bioinformatics or phylogenetic methods. If in Cryptococcus 18S rRNA cytosines are modified like in S. cerevisiae the function to guide Kre33/NAT10 to its substrate might be covered by other molecules. Secondary structures (drawn with R2R) and speculative U13-like interactions with the 18S rRNA 3’ end can be modeled for Tremellomycetes U3/snR45 (bottom row):
Tremellomycetes U3 Close homologues of S. cerevisiae U3 Yeast U3 model
Samarsky et al 1998 at www.ResearchGate.net
![From Samarsky et al. 1998: The U3 snoRNA from the yeast S. cerevisiae. The model shown is derived from a compilation of structures proposed for U3 RNAs from different organisms. Several configurations have been suggested for the 5 segment including (i) nonfolded, (ii) possessing a single helix (hairpin 1), or (iii) containing two helices (hairpin 1a and 1b, not shown). The well-structured 3' portion of S. cerevisiae U3 contains two highly conserved (central and terminal stems) and nonconserved helices (hairpins 2, 3, and 4). The number of hairpins in this region varies from none to three in different organisms. The relative positions of the phylogenetically conserved sequence elements, boxes A, A', B, C, C', and D are well preserved in all U3 snoRNAs known. The box A and A' segments are believed to interact with pre-rRNA (see text). Box C influences association with the protein fibrillarin (6). Box D is required for formation of the 5' TMG cap structure and nuclear retention of mature RNA. The primary sequence of the hinge region separating the conserved elements of the 5' and 3' portions is not well preserved phylogenetically. Ten nucleotides of the hinge segment in yeast U3 have been postulated to interact with the 5' ETS of pre-rRNA through complementary base-pairing. The U3 snoRNA from the yeast S. cerevisiae. The model shown is derived from a compilation of structures proposed for U3 RNAs from different organisms. Several configurations have been suggested for the 5' segment including (i) nonfolded (28, 49), (ii) possessing a single helix (hairpin 1 [50]), or (iii) containing two helices (hairpin 1a and 1b, not shown [44, 52]). The well-structured 3' portion of S. cerevisiae U3 contains two highly conserved (central and terminal stems) and nonconserved helices (hairpins 2, 3, and 4 [26]). The number of hairpins in this region varies from none to three in different organisms. The relative positions of the phylogenetically conserved sequence elements, boxes A, A', B, C, C', and D are well preserved in all U3 snoRNAs known. The box A and A' segments are believed to interact with pre-rRNA (see text). Box C influences association with the protein fibrillarin (6). Box D is required for formation of the 5' TMG cap structure and nuclear retention of mature RNA (68). The primary sequence of the hinge region separating the conserved elements of the 5' and 3' portions is not well preserved phylogenetically. Ten nucleotides of the hinge segment in yeast U3 have been postulated to interact with the 5' ETS of pre-rRNA through complementary base-pairing (8).](https://www.researchgate.net/profile/Dmitry-Samarsky/publication/13700275/figure/fig8/AS:668739427135491@1536451312152/The-U3-snoRNA-from-the-yeast-S-cerevisiae-The-model-shown-is-derived-from-a-compilation.png)
Tremellomycetes U3 as snR45 U13 (mammal, plant) Tremellomycetes snR45-II
The insets show proposed base pairing between the 3’ region of 18S rRNA and yeast snR45 or human U13 (from [Sharma-2017]). Helix-formation by association of C/D and C’/D’ motifs has been incorporated in these models (in contrast to the U3 models in the top row). All U3 interaction sites around the acetylated C near the 3’ end of 18S rRNA, as found by snR45/U13, differ from those that sustain the pseudo-loop.
The modeled interactions between Tremellomycetes U3 snoRNA and 18S rRNA cover half of the overall base pairing interactions that can be formed between snR45/U13 and 18S rRNA nearby the acetylated cytosine. A second conserved element in snR45/U13 forms a helix with 18S rRNA nucleotides upstream of the modified cytosine (C1773 in 18S rRNA of S. cerevisiae). Sequence conservation that would fully support such a helical structure in the case of Tremellomycetes is not observed.
A snoRNA specific for Tremellomycetes (snR45-II), however, is identified that could provide these contacts. Alternatively, this snoRNA might regulate the putative base-pairing of U3 with the 3’ end of 18S rRNA. The activity of this snoRNA appears to be controlled; snR45-II can adopt different conformations mediated by rearrangement of base-pair interactions between three stretches of well-conserved nucleotide sequences (see here).
Therefore, the functions of yeast snR45 and U3 could have been distributed over several snoRNAs in Tremellomycetes. Confirmation of acetylation of cytosines in 18S rRNA followed by crosslinking experiments to isolate RNAs binding to Kre33/Nat10 from Cryptococcus might resolve this; still, a complication could be that U3 was a common contaminant in such experiments in baker’s yeast [Sharma-2017].
The Cryptococcus U3 snoRNAs and snR45-II are further described in: snR45-U3a, U3b, and snR45-II.