U14 & snR190¶
In yeast S. cereviaiae the snoRNA snR128, or U14, is essential for maturation of the small ribosomal subunit containing the 18S rRNA. It is transcribed as part of a bicistronic precursor [Zagorski-1988], also containing snoRNA snR190, involved in compaction of the 60S large subunit precursor, together with Npa1 and Dbp7 [Aquino-2021, Jaafar-2021]. The snR190-U14 containing transcript is processed by Rnt1, Xrn1 and Rat1 [Chanfreau-1998; Petfalski-1998].
Both snoRNAs contain relevant stretches of complementarity with rRNA. Apart from modification guides, many snoRNAs harbor conserved sequences that support additional base pairing [van.Nues-2011], one of which, in U14, is within an essential structural element, the Y-domain [Samarsky-1996].
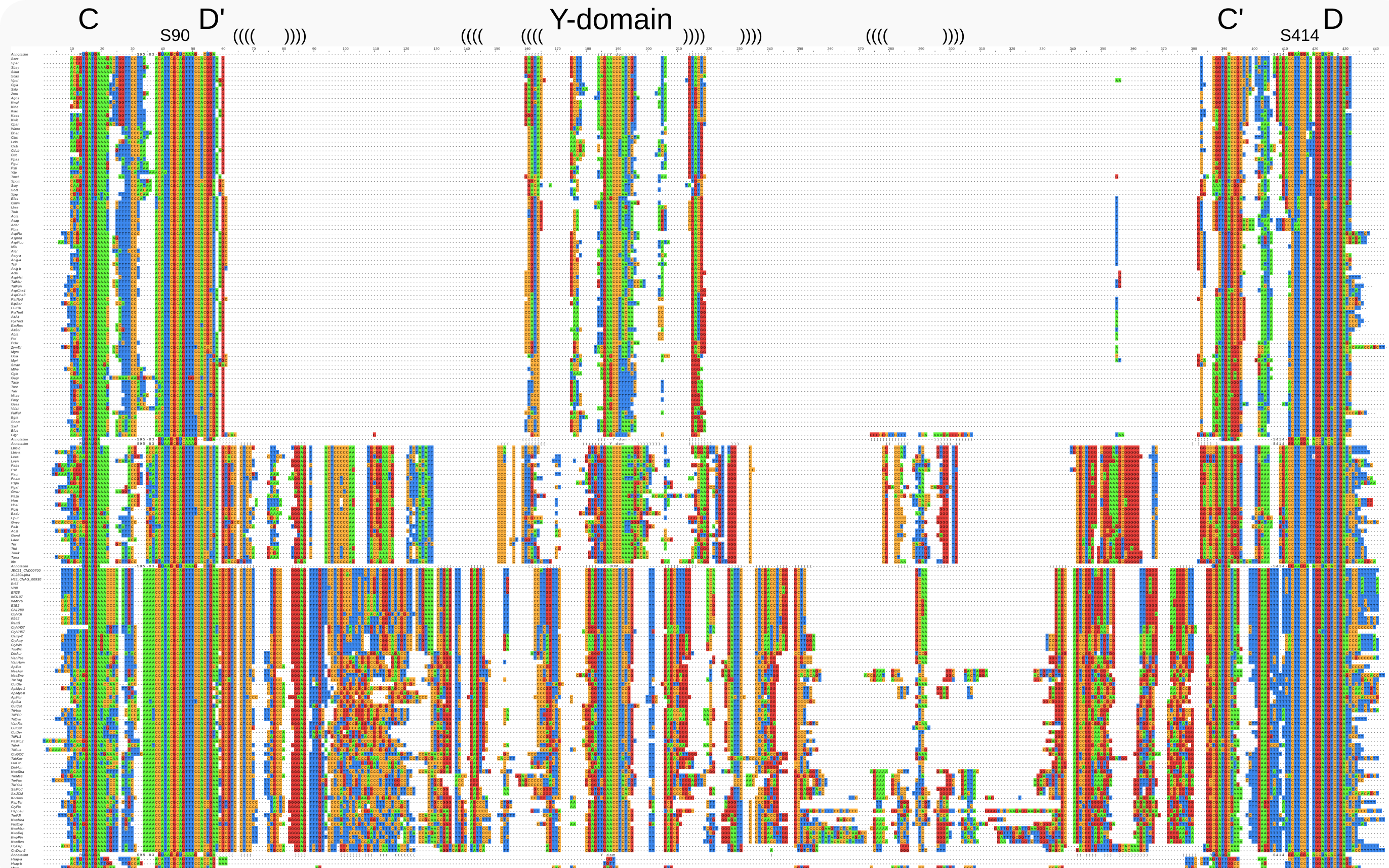
Alignments for fungal U14 snoRNAs¶
In other fungi, the U14 sequences contain similar regions of homology including the Y-domain, which also can be discerned in the U14 snoRNA of Cryptococcus.
In jelly fungi (Tremellales), U14 is encoded in the 2nd 5’ intron of the gene for Argininosuccinate-synthase (CND00700 in C. deneoformans JEC21 or CNAG_00930 in C. neoformans H99). The first intron of this gene also encodes a snoRNA which is the homologue of snR190 as based on conservation of structural elements that may provide extra base pairing [van.Nues-2011]. The primary sequence of snR190 in yeast is much longer than in Pezizomycotina [van.Nues-2011] or Tremellomycetes, while this is the case for U14 in Tremellomycetes.
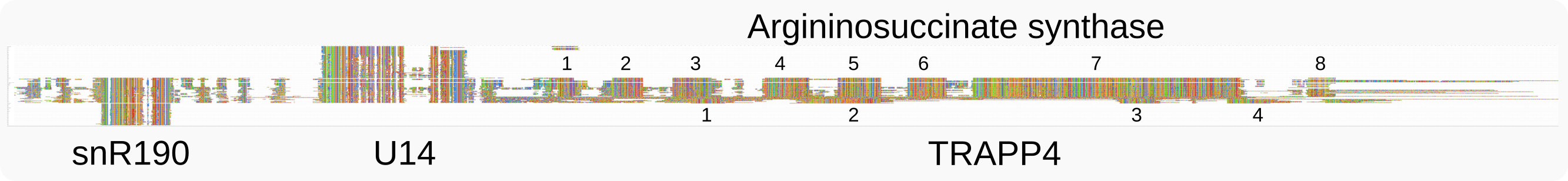
The snR190 and U14 snoRNAs in Tremellales are part of the transcript for Argininosuccinate-synthase¶
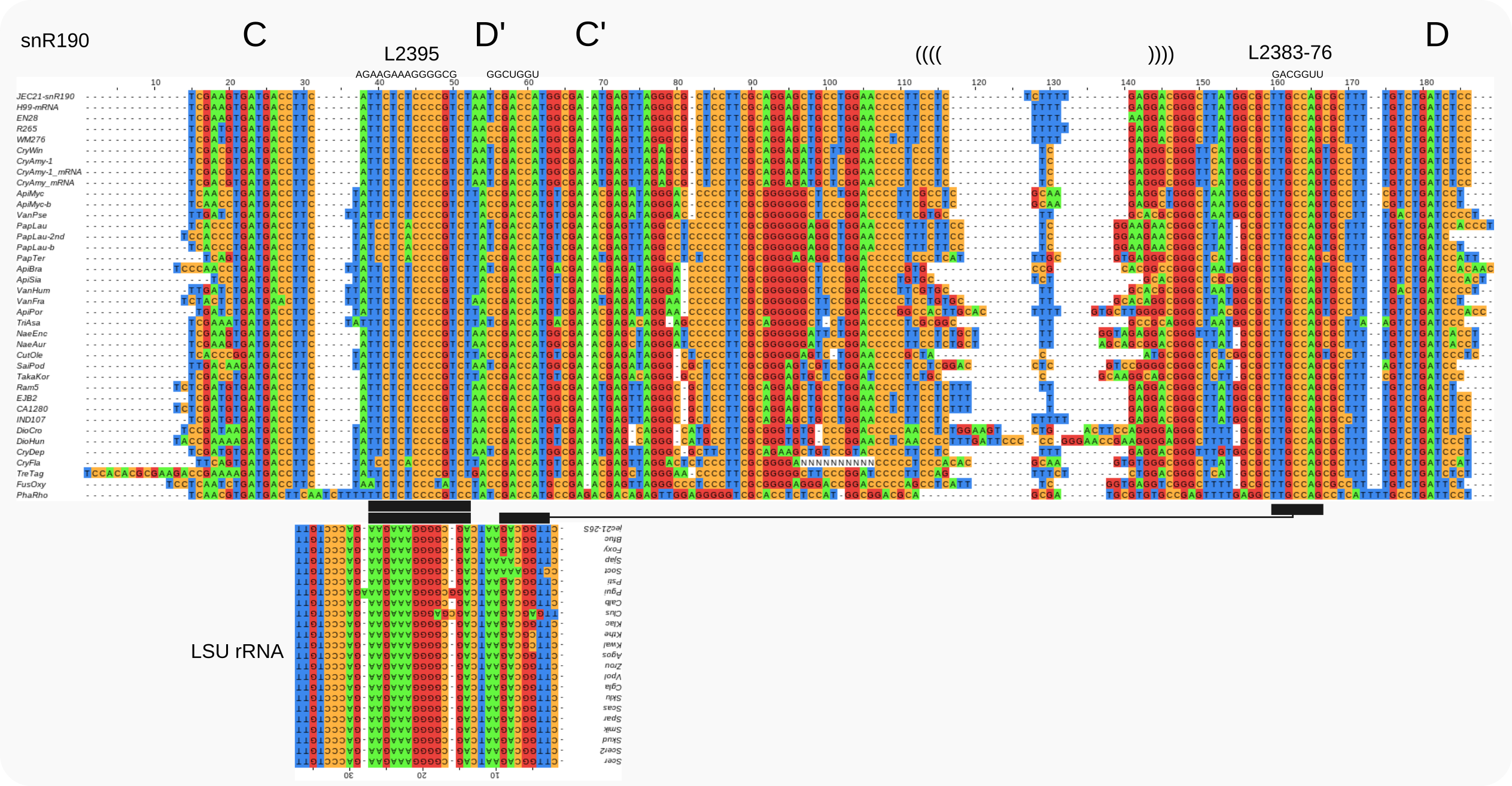
Alignments for snR190 snoRNAs in Tremellomycetes¶
Large subunit rRNA target regions for conserved regions in snR190 are shown below the snoRNA alignment
The linkage and conserved 5’ to 3’ order of the genes for snR190 and U14 is observed in various classes of fungi. Compared to jelly fungi, both snoRNA genes are connected to different genes in more distant Tremellomycetes:
Apiotrichum porosum and Vanrija; ~440-500 nt downstream of U14 is the ATG of TRAPP4, a core component of the transport protein particle complex and one of the essential subunits for guanine nucleotide exchange factor (GEF) activity for Rab1 GTPase (like CNAG_00931 or CND00710).
Trichosporon asahii var. asahii; the gene for a hypotheical protein with similarity to PPP1R42 (protein phosphatase 1 regulatory subunit 42; like CNAG_03685 or CNB01830).
Cryptococcus depauperatus; ~350 nt downstream of U14 is the ATG of the gene for a CCD97-like C-terminal domain-containing protein (like CNAG_00892 or CND00310).
Apiotrichum brassicae; the gene for ATP-dependent RNA helicase DSR1 (like DDX27, homologous to CNAG_01140 or CND02650), involved in 60S maturation, but transcribed from the opposite strand.
Apiotrichum siamense; again DSR1 but on the coding strand, the ATG is 540 nt downstream of U14.
Although the organization of both snoRNA genes might suggest that fungal ribosome synthesis relies on equimolar levels and synchronous expression of U14 and snR190, these snoRNAs are expressed from separate loci in some Pezizomycotina such as Aspergillus or Talaromyces species.
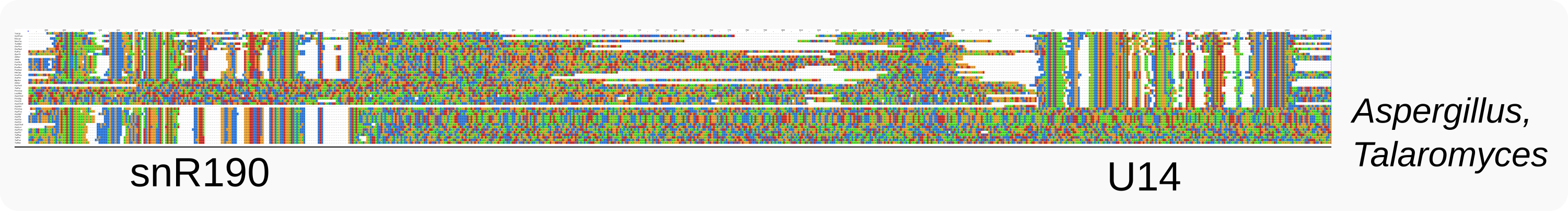
Genes for U14 and snR190 are uncoupled in some Pezizomycotina, like Aspergillus and Talaromyces¶
U14-SNORD14A¶
U14_intronic-boxCDsnoRNA-258nt
Within 2 nd intron of 5UTR of CND00700 for Argininosuccinate-synthase
Predicted target in 18S rRNA: acatCcaaggaagg
Orthologue of yeast U14 , human SNORD14A (U14A) or SNORD14B (U14B)
>AE017344.1:195237-195495 Cryptococcus neoformans var. neoformans JEC21 chromosome 4 sequence TTTCTATGATGAAACCCAATGTAAAACCATACGCAGTTTCCACTGAACGCGTCCTCCTTGCCGGGAGTTT GTTCCTCGCCCTTTTCTCCTCGGTCTCGCTTCTGAAACTGAGTTGAGTCTTCCATGGTTCCGAGTTGAAC CTCTCGTTGAGCTTTGGACAGATTCCTCGACCTCGGGCTCGTAAGAGCAGTCGGTACGATCGTTGGGGAA GGGCTTGGCGATGCTAGTTTGAAGTTTTTTCTTCCTTGGATGTCTGATT
snR190-SNORD12B¶
snR190_intronic-boxCDsnoRNA-145nt
Within 1 st intron of 5UTR of CND00700 for Argininosuccinate-synthase
Predicted target in 25S rRNA: gcggGgaaagaa
In fission yeast the conserved element GCCAGT [van.Nues-2011] is part of a sequence that “also mediates the Mmi1-Mei2 mutual control in vegetative cells and restricts meiotic gene expression during sexual differentiation” [Leroy-2025]; the conserved loop [van.Nues-2011] connecting boxes D’ and C’ is required for guiding rRNA 2’-O-methylation and “scaffolding gametogenesis effectors” [Leroy-2025].
Orthologue of yeast snR190, human HBII-99B (SNORD12B), SNORD12, SNORD12C, or fission yeast snR107
>AE017344.1:194962-195105 Cryptococcus neoformans var. neoformans JEC21 chromosome 4 sequence TCGAAGTGATGACCTTCATTCTCTCCCCGTCTAATCGACCATGGCGAATGAGTTAGGGCGCTCCTTCGCA GGAGCTGCCTGGAACCCCTTCCTCTCTTTTGAGGACGGGCTTATGGCGCTTGCCAGCGCTTTTGTCTGAT CTCC
Image source U14:
# STOCKHOLM 1.0
#=GF Comparative analysis of Tremellomycetes U14 snoRNA (in context of other U14 snoRNAs)
#=GF RW van Nues, May 2023
#=GF https://coalispr.codeberg.page/paper
#=GF https://doi.org/10.1038/emboj.2011.148
Scer/1-122 ---------ACGGTGATGAAAGACTGGTTCCTTA---ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CTT-----ACGAACCCATCGT--------TA-------GTACTC----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CGGTGACCGCTCT-TCTTT-AGAGACCTTCCTA-GGATGTCTGAGT-------------
#=GS SacCer/1-126 DE NC_001142.9:c139691-139566 Saccharomyces cerevisiae S288C chromosome X, complete sequence
SacCer/1-126 -------TCACGGTGATGAAAGACTGGTTCCTTA---ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CTT-----ACGAACCCATCGT--------TA-------GTACTC----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CGGTGACCGCTCT-TCTTT-AGAGACCTTCCTA-GGATGTCTGAGTGA-----------
Spar/1-122 ---------ACGGTGATGAAAAACTGGTTCCTTA---ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CTT-----ACGAACCCATCGT--------TA-------GTACTC----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CGGTGACCGCTCT-TCTAT-AGAGACCTTCCTA-GGATGTCTGAGT-------------
Sbay/1-123 ---------ACAGTGATGAAAGACTGGTTCCTTGA--ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CTT-----ACGAACCCATCGT--------TA-------GTACTC----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CGGTGACCGCTCT-ATTTT-AGAGACCTTCCTA-GGATGTCTGAGT-------------
Skud/1-122 ---------ACAGTGATGAAAAACTGGTTCCTTT---ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CTT-----ACGAACCCATCGT--------TA-------GTACTC----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CGGTGACCGCTCT-TCTAT-AGAGACCTTCCTA-GGATGTCTGAGT-------------
Scas/1-121 ---------ACGATGATGAAAA-TTGGTTCCTTA---ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------TGGTAC---G----CTT-----AAGAACCCATCTT--------TA-------GTACCA----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CGGTGACCGCTCA-TTTAT-TGAGACCTTCCTA-GGATGTCTGAGT-------------
Vpol/1-120 ---------ACGATGATGAAAA-TCGGTTCCTT----ACATTCGCAGTTTCCACGGTA-G-----------------------------------------------G-----------------------------------------------------AGTACG-------CTT-----ACGAACCCATCGT--------T-------TGTACT----------------------------------------------------------------------------------------------------------------------------------------AA-----------------------T------CGGTGACCGCCTC-TTTAT--GAGACCTTCCTA-GGATGTCTGAGT-------------
Cgla/1-122 ---------ACGATGATGAAAATTCGGTTCCTT----ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CCTT----ACGAACCCATCGT--------TA-------GTACTT----------------------------------------------------------------------------------------------------------------------------------------------------------------TT-----CGGTGACCGCCTC-TTTAC--GAGACCTTCCTA-GGATGTCTGAGT-------------
Sklu/1-123 ---------AAGGTGATGAAAATCTGGTTCCTT----ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGCAC---G----CTTAA---ACGAACCCATCGT-------ATA-------GTGCTC----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CGGTGACCGCTCTC-TTAC--GAGACCTTCCTA-GGATGTCTGATT-------------
Zrou/1-123 ---------ACTATGATGAAACTTTGGTTCCTTGA--ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CCTT----TCGAACCCATCGA------TATA-------GTGCTC----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CGGTGACCGCTCA--TT---TGAGACCTTCCTA-GGATGTCTGAGT-------------
#=GS ZygRou/1-131 DE NC_012993.1:c171721-171591 Zygosaccharomyces rouxii strain CBS732 chromosome D complete sequence
ZygRou/1-131 -----GATCACTATGATGAAACTTTGGTTCCTTGA--ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CCTT----TCGAACCCATCGA------TATA-------GTGCTC----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CGGTGACCGCTCA--TT---TGAGACCTTCCTA-GGATGTCTGAGTGATC---------
Agos/1-119 ---------AAGGTGATGAAAA-TTGGTTCCTTA---ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----C------TTCGAACCCATCGTA-------TA-------GTGCTC----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CGGTGACCGCTC--TTATT--GAGACCTTCCTA-GGATGTCTGATT-------------
Kwal/1-119 ----------CGATGATGAAAATCTGGTTCCTT----ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGCAC---G----CCA-----ACGAACCCATCGT-------ATA-------GTGCTC----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CGGTGACCGCTC--TTCTT--GAGACCTTCCTA-GGATGTCTGAGT-------------
Kthe/1-120 ---------GCGATGATGAAAACTTGGTTCCTT----ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CCA-----ACGAACCCATCGT-------ATA-------GTGCTC----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CGGTGACCGCTC--TTCTT--GAGACCTTCCTA-GGATGTCTGAGT-------------
#=GS KluLac/24-149 DE NC_006037.1:555022-555187 Kluyveromyces lactis strain NRRL Y-1140 chromosome A complete sequence
KluLac/24-149 ---------TATATGATGAAAA-TTGGTTCCTTT---ACATTCGCAGTTTCCACGGT-AG----------------------------------------------------------------------------------------------------GAGTAC---G----CCT-----ACGAACCCATCGT--------TA-------GTACTC----------------------------------------------------------------------------------------------------------------------------------------------------------------TT-----CAGTGACCGCTC--TTTATT-GAGACCTTCCTA-GGATGTCTGATCTTTTT--------
Kaes/1-121 ---------TATATGATGAAAG-TTGGTTCCTTT---ACATTCGCAGTTTCCACGGT-AG----------------------------------------------------------------------------------------------------GGGTAC---G----CCT-----ACGAACCCATCGT--------TA-------GTACTC----------------------------------------------------------------------------------------------------------------------------------------------------------------TT-----CAGTGACCGCTC--TTAATT-GAGACCTTCCTA-GGATGTCTGATT-------------
Kwic/1-120 ---------TAGATGATGAAAATTTGGTTCCTTT---ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CTT-----ACGAACCCATCGT--------TA-------GTACCC----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CAGTGACCGCTC--TTTAT--GAGACCTTCCTA-GGATGTCTGATT-------------
Cpar/1-116 ---------AAGGTGATGAAAGACTGGTTCCTTA---ACATTCGCAGTTTCCACGGTA-G----------------------------------------------------------------------------------------------------GAGTAC---G----CTT-----ACGAACCCATCTT--------T--------GTATGT----------------------------------------------------------------------------------------------------------------------------------------------------------------C-------AGTGACCGTCG----AAA---TGACCTTCCTTTGGATGTCTGATT-------------
Wano/1-108 ---------AAGATGATGAAAC----TATCCATA---ACATTCGCAGTTTCCACGGTA-G-----------------------------------------------------------------------------------------------------CATAC---G----TAC-----AAGAACCCATCTT---------A-------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CAGTGACCGT-----TAAT----GTACTTCCTT-GGATGTCTGATT-------------
Dhan/1-109 ---------TATATGATGAAAA----TTTCCCATTA-ACATTCGCAGTTTCCTCGGTA-G-----------------------------------------------------------------------------------------------------CATAC---A----CT-------AGAACCCATCT---------TC-------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CGGTGACCGCT-----TAT---AGACCTTCC-ATGGATGTCTGATA-------------
Clus/1-107 ---------TAAGTGATGAAAT-----ATCCCATA--ACATTCGCAGTTTCCTCGGTA-G-----------------------------------------------------------------------------------------------------CATAC---A----A-------TAGAACCCAATCTA----------------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CGGTGACCGC----TTTAT----GACCTTCC-ATGGATGTCTGATT-------------
Lelo/1-112 ---------AAGGTGATGAAAA---CGTACCATA---ACATTCGCAGTTTCCTCGGTA-G-----------------------------------------------------------------------------------------------------CATAC---A----ACAC------GAACCTAATTC-------AT--------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------T------CGGTGACCGCCG----AAT---CGACCTTCCTTTGGATGTCTGATT-------------
Calb/1-115 ---------AAGGTGATGAAAA--ATTTTCCCAA---ACATTCGCAGTTTCCTCGGTA-G-----------------------------------------------------------------------------------------------------CATAC---A----ACGA----C-GAACCTAATC--------ACA-------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CGGTGACCGC----TCCATAC--GACCTTCCTTTGGATGTCTGATT-------------
Cdub/1-116 ---------AAGGTGATGAAAA--GTTTTCCCAA---ACATTCGCAGTTTCCTCGGT-AG-----------------------------------------------------------------------------------------------------CATAC---A----ACGA----C-GAACCTAATC--------TCA-------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------TT-----CGGTGACCGC----TCTATAC--GACCTTCCTTTGGATGTCTGATT-------------
Ctro/1-110 ------------GTGATGAAAT--TTTTTCCCAA---ACATTCGCAGTTTCCTCGGTA-G-----------------------------------------------------------------------------------------------------CATAC---G----ACAC------GAACCTAATC-------TACT-------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CGGTGACCGCT-----TAT---AGACCTTCCATTGGATGTCTGATT-------------
Ppas/1-112 ---------TACATGATGAAAT--CTATTTCTAT---ACATTCGCAGTTTCCTCGGTA-G-----------------------------------------------------------------------------------------------------CATAC---G----CAC-----AAGAACCCATCTT---------T-------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CGGTGACCGCC----ACATA---GACCTTCCT-TGGATGTCTGATT-------------
Pgui/1-107 ---------TATATGATGAAAG----ATTCCATAA--ACATTCGCAGTTTCCTCGGTA-G-----------------------------------------------------------------------------------------------------CATAC---G----C--------AGAACCCATCT---------TA-------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CAGTGACCGC-----TTAAT---GACCTTCCT-TGGATGTCTGATA-------------
Psti/1-108 ---------AAAGTGATGAAAG----TTACCATAA--ACATTCGCAGTTTCCTCGGTA-G-----------------------------------------------------------------------------------------------------CATAC---A----AT-------AGAACCCATCT---------TA-------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CGGTGACCGC-----TTATC---GACCTTCCT-TGGATGTCTGATT-------------
Ylip/1-110 ---------TTTCTGATGAAAT----TTTCATTTTAAACAATCGCAGTTTCCTCGGTA-G-----------------------------------------------------------------------------------------------------CATAC---G----TA------TAGAACCCATCTA-----------------GTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------C------CAGTGACCGTT----GTAT----GACCTTTCT-TGGATGTCTGATT-------------
Tmel/1-112 ---------ACCATGATGAAAA---AATTCCT-----ACATTCGCAGTTTCCTCGGT-A-----------------------------------------------------------------------------------------------------GCACAC---G----C-------TAGAACCCATTCTA------TAA-------GTGTGC---------------------------------------------------------------------------------------------------------------------------------------G-------------------------T--A--ACGTGACCGCT----GCA----AGACCTTCCT-TGGAAGTCTGATT-------------
Spom/1-106 ---------CAGGTGATGAAAT-----TTCCATTGA-ACATTCGCAGTTTCCCCGGA-GC-----------------------------------------------------------------------------------------------------GGCA----T----AC--------GAACCCAATT--------T----------TGCC-----------------------------------------------------------------------------------------------------------------------------------------------------------------GC-----AGATGTCGTGC-----ATA----GTCCTTCCTTTGGATGTCTGATG*------------
Scry/1-105 ---------CAAGTGATGAAAT-----TTCCAATAA-ACATTCGCAGTTTCCACGGA-GC-----------------------------------------------------------------------------------------------------GACATA--------C--------GAACCCATTC-----------------TA-GTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------GC-----AAATGACGGC-----CATA----GTCCTTCCTTTGGATGTCTGATG-------------
Soct/1-105 ---------CAGGTGATGAAAT-----TTCCAACAA-ACATTCGCAGTTTCCACGGA-GC-----------------------------------------------------------------------------------------------------GACA----T----AC--------GAACCCATTC--------T----------TGTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------GC-----AAATGACGGC-----CATA----GTCCTTCCTTTGGATGTCTGATG-------------
Sjap/1-105 ---------CGTGTGATGATAA---TTTTCCACAA--ACATTCGCAGTTTCCACGGA-TC-----------------------------------------------------------------------------------------------------GACA----T----A--------AGAACCCAATCT------------------TGTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------GG-----ATATGACGGC-----CCA-----GTCCTTCCTTTGGATGTCTGATG-------------
Efes/1-104 ---------CATATGATTATAT----TTTCCCAT---TAATTCGCAGTTTCCACGCTA-GC-----------------------------------------------------------------------------------------------------CGTC--------TT-------AGAGCCTTT---------------------GACG----------------------------------------------------------------------------------------------------------------------------------------T-----------------------GC-------AGTGAGCGAT---TGAAT---ATGCCTACCT-TGGATGTATGATG-------------
Cimm/1-107 ---------TTTATGATGAAAC--CTTTTCCT-----TCATTCGCAGTTTCCACGCTA-GC----------------------------------------------------------------------------------------------------CGTC----G-----------CATGAACCTATTAAG------TC---------GACG----------------------------------------------------------------------------------------------------------------------------------------T-----------------------GT------CGTTGAGCGC------ATT-----GCCTACCT-TGGACGTCTGAGA-------------
Uree/1-107 ---------CTCATGATGAAAC--CTTTTCCT-----TCATTCGCAGTTTCCACGCTA-GC----------------------------------------------------------------------------------------------------CGTCG--------TA-------TGAACCTAGTA--------AAC-------CGACG----------------------------------------------------------------------------------------------------------------------------------------T-----------------------GT------CGTTGAGCGC------ATT-----GCCTACCT-TGGACGTCTGATT-------------
Trub/1-107 ---------TCTATGATGAAAT--TTTTTCCT-----ACATTCGCAGTTTCCACGCTA-GC----------------------------------------------------------------------------------------------------TGTCG--------CA-------TGAACCTAATA--------AGT-------CGACG----------------------------------------------------------------------------------------------------------------------------------------T-----------------------GT------CGTTGAGCGT------TTT-----ATCTTCCT-TGGATGTCTGATT-------------
Aota/1-107 ---------TCTATGATGAAAT--TTTTTCCT-----ACATTCGCAGTTTCCACGCTA-GC----------------------------------------------------------------------------------------------------TGTCG--------CA-------TGAACCTATTA--------AGT-------CGACG----------------------------------------------------------------------------------------------------------------------------------------T-----------------------GT------CGTTGAGCGG-----ATTA------TCTTCCT-TGGATGTCTGATT-------------
Mcan/1-104 -----------TATGATGAAAT--TTTTTCCT-----ACATTCGCAGTTTCCACGCTA-GC----------------------------------------------------------------------------------------------------TGTCG--------CA-------TGAACCTATTA--------AGT-------CGACG----------------------------------------------------------------------------------------------------------------------------------------T-----------------------GT------CGTTGAGCGG-----ATTA------TCTTCCT-TGGATGTCTGAT--------------
Acap/1-116 ---------CGTATGATGAAAT--TTTTTCCC-----ACATTCGCAGTTTCCACGCTA-GC----------------------------------------------------------------------------------------------------CGTCG--------CA-------TGAACCTATTA--------AGT-------CGACG----------------------------------------------------------------------------------------------------------------------------------------T-----------------------GT------CGTTGAGCGGACAA-TAAAT-TTGCCTTACCT-TGGATGTCTGATC-------------
Ader/1-115 ---------CTCATGATGAAAT--TTTTTCCT-----ACATTCGCAGTTTCCACGCTA-GC----------------------------------------------------------------------------------------------------CGTCG--------CA-------TGAACCTATTA--------AGT-------CGACG----------------------------------------------------------------------------------------------------------------------------------------T-----------------------GT------CGTTGAGCGGACAA-TTAA--TTGCCTAACCT-TGGATGTCTGATT-------------
Pbra/1-114 ---------CTCATGATGAAAT--TTTTTCCT-----ACATTCGCAGTTTCCACGCTA-GC----------------------------------------------------------------------------------------------------CGTCG--------CA-------TGAACCTAATA--------AGT-------CGACG----------------------------------------------------------------------------------------------------------------------------------------T-----------------------GT------CGTTGAGCGGACAA--ATA--TTGCCTAACCT-TGGATGTCTGATT-------------
Afla/1-103 ---------TCGATGATGAAAA--TTTTTCCC-----ACATTCGCAGTTTCCACGCT-AGC----------------------------------------------------------------------------------------------------CGTC----G----C-------TAGAACCCAATCTA-----------------GACG----------------------------------------------------------------------------------------------------------------------------------------T-------------------------GCT-----CTGTGCGC------AAT------ACTTCCT-TGGATGTCTGAGA-------------
Anid/1-104 ---------TCGATGATGAAAA-AGTTTTCC------ACATTCGCAGTTTCCACGCT-AGC----------------------------------------------------------------------------------------------------CGTC----G----C-------TAGAACCCAATCTA-----------------GACG----------------------------------------------------------------------------------------------------------------------------------------T-------------------------GCT-----CTGTGCGC-----ATGT------ACTTCCT-TGGATGTCTGACG-------------
Aory-b/1-103 ---------TCGATGATGAAAA--TTTTTCCC-----ACATTCGCAGTTTCCACGCT-AGC----------------------------------------------------------------------------------------------------CGTC----G----C-------TAGAACCCAATCTA-----------------GACG----------------------------------------------------------------------------------------------------------------------------------------T-------------------------GCT-----CTGTGCGC------AAT------ACTTCCT-TGGATGTCTGAGA-------------
Nfis/1-102 ---------TAAATGATGAAAA-CCTTTTCC------ACATTCGCAGTTTCCACGCT-AGC----------------------------------------------------------------------------------------------------CGTC----G----CT-------AGAACCCATCT--------TA---------GACG----------------------------------------------------------------------------------------------------------------------------------------T-------------------------GCT-----CTGTGCGC-----ATTT------TCTTCCT-TGGATGTCTGA---------------
Ater/1-101 -----------TATGATGAAAA-TTATTCCCT-----ACATTCGCAGTTTCCACGCT-AGC----------------------------------------------------------------------------------------------------CGTC----G----CC------ATGAACCCATCAT--------A---------GACG----------------------------------------------------------------------------------------------------------------------------------------T-------------------------GCT-----CTGTGCGC------AAT------ACTTCCT-TGGATGTCTGA---------------
Aory-a/1-104 ---------TTTATGATGAAAA--ATTTTCCC-----ACATTCGCAGTTTCCACGCT-AGC----------------------------------------------------------------------------------------------------CGTC----G----C-T-----ATGAACCTATA---------TCA--------GACG----------------------------------------------------------------------------------------------------------------------------------------T-------------------------GCT-----CTGTGCGC------AAT------ACTTCCT-TGGATGTCTGACA-------------
Anig-a/1-103 ---------TCGATGATGAAAA--CTTTTCCT-----ACATTCGCAGTTTCCACGCT-AGC----------------------------------------------------------------------------------------------------CGTC----G----CG-------AGAACCCATTCT--------A---------GACG----------------------------------------------------------------------------------------------------------------------------------------T-------------------------GCT-----CTGTGCGC------AAT------ACTTCCT-TGGATGTCTGACG-------------
Tsti/1-107 ---------TTTATGATGAAAA-CATTTTCC------ACATTCGCAGTTTCCACGCT-AGC----------------------------------------------------------------------------------------------------CGTC----G----CC------GTGAACCCAATTCC------ATA--------GACG----------------------------------------------------------------------------------------------------------------------------------------T-------------------------GCT-----CTGTGCGC------ATT------ACTTCCT-TGGATGTCTGATC-------------
Acla/1-101 -----------TATGATGAAAA--CTTTTCCT-----ACATTCGCAGTTTCCACGCT-AGC----------------------------------------------------------------------------------------------------CGTC----G----CC------ATGAACCCATCAT--------A---------GACG----------------------------------------------------------------------------------------------------------------------------------------G-------------------------GCT-----CTGTGCGC-----ATTA------TCTTCCT-TGGATGTCTGA---------------
Pnod/1-103 ---------TTCATGATGAAAC---ATTTCC------ACATTCGCAGTTTCCACGCTA-GC----------------------------------------------------------------------------------------------------CATC---------AA------TTGAACCTACAA--------CC---------GATG----------------------------------------------------------------------------------------------------------------------------------------T-------------------------GC-----CATGAGCGC-----GAATA-----CCTTCCT-TGGATGTCTGATC-------------
Anig-b/1-103 ---------CTTATGATGAAAA--CTTTTCCT-----ACATTCGCAGTTTCCACGCT-AG----------------------------------------------------------------------------------------------------TCGTC----G----CC------ATGAACCCATCAT--------A---------GACGG------------------------------------------------------------------------------------------------------------------------------------------------------------------CT-----CTGTGCGC------AAT------ACTTCCT-TGGATGTCTGACT-------------
Pchr/1-102 ---------TCGATGATGAAAC---TCTTCCT-----ACATTCGCAGTTTCCACGCTA-G----------------------------------------------------------------------------------------------------CCGTC---------GC------TTGAACCCAATCAA-----------------GGCGG------------------------------------------------------------------------------------------------------------------------------------------------------------------C-----ATTGAGCGG-----AATA------TCTTCCT-TGGATGTCTGAGA-------------
Abra/1-103 ---------TTCATGATGAAAC---ATTTCC------ACATTCGCAGTTTCCACGCTA-G----------------------------------------------------------------------------------------------------CCATC---------AA------TTGAACCTACAA--------CC---------GATGG---------------------------------------------------------------------------------------------------------------------------------------A--------------------------C-----AATGAGCGC-----GAATA-----CCTTCCT-TGGATGTCTGATC-------------
Ptri/1-103 ---------TACATGATGAAAC---ATTTCC------ACATTCGCAGTTTCCACGCTA-G----------------------------------------------------------------------------------------------------CCATC---------AA------TTGAACCTACAA--------CC---------GATGG---------------------------------------------------------------------------------------------------------------------------------------A--------------------------C-----AATGAGCGC-----GAATA-----CCTTCCT-TGGATGTCTGATA-------------
Mgra/1-103 ---------TGGATGATGAAAA-ACTTTTCC------ACATTCGCAGTTTCCACGCTA-G----------------------------------------------------------------------------------------------------CCGTC---------GC-----TACGAACCTAATA-------------------GACGG---------------------------------------------------------------------------------------------------------------------------------------A--------------------------C-----GATGAGGGC-----CCAA------TCTTCCT-TGGATGTCTGACA-------------
Gcla/1-107 ---------TGCATGATGAAAC--ATTTTCCT-----ACATTCGCAGTTTCCACTTGA-GC----------------------------------------------------------------------------------------------------ATCC---------ATC------AGAGCCTAATCT--------ACC-------GGAT----------------------------------------------------------------------------------------------------------------------------------------C-------------------------GC----ATATGAGGGC-----GAAT------TCTTCCT-TGGATGTCTGATC-------------
Mgri/1-104 ---------TTTATGATGAAAC---ATTTCCT-----ACATTCGCAGTTTCCACTCTATGC-----------------------------------------------------------------------------------------------------CCC----G----TCA-------GAACCTTTC----------A---------GGG-----------------------------------------------------------------------------------------------------------------------------------------AT------------------------GCA----AATGAGAGC----GAATAA-----CCTTCCT-TGGATGTCTGATC-------------
Smac/1-103 ---------CTTATGATGAAGA----TTTCCT-----ACATTCGCAGTTTCCACTCGAGAC-----------------------------------------------------------------------------------------------------CCC----A----CCT-----ACGAGCCTTTCTC------------------GGG-------------------------------------------------------------------------------------------------------------------------------------------------------------------GTC---AGATGAGGGC-----GAAG------TCTTCCT-TGGATGTCTGATT-------------
#=GS Mthe/1-104 DE CP003008.1:1507881-1507984 Myceliophthora thermophila ATCC 42464 chromosome 7, complete sequence
Mthe/1-104 -------TCCATATGATGAAAT----TTTCCCAT---ACATTCGCAGTTTCCACTCGA-G-----------------------------------------------------------------------------------------------------TCCC----A----CC-------AGAGCCTATCTC------------------GGGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGGGC-----GAAT-----ACCTTCCT-TGGATGTCTGATT-------------
Cglo/1-99 ---------CGTATGATGAAAT----TTTCCCT----ACATTCGCAGTTTCCACTCGA-G------------------------------------------------------------------------------------------------------TCC----T----AAA------AGAGCCTTTTCT------------------GGA--------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AAATGAGAGC-----GAAT------CCTTCCT-TGGATGTCTGACG-------------
#=GS Gagr/1-106 DE Gaeumannomyces graminis var. tritici R3-111a-1; Gae_graminis_V2 supercont2.2: 5,312,077-5,312,194
Gagr/1-106 ---------AAAATGATGAAT3TCCAAACAAGTGCCTACATTCGCAGTGGCCTCTCGA-G------------------------------------------------------------------------------------------------------CCC----T----T--------AGAGCCTTTTTT------------------GGG--------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGAGC-----AAATA-----CCTTCCT-TGGATGTCTGATT-------------
Tasp/1-101 ---------TGCATGATGAAAA----TTTCCATT---TAATTCGCAGTTTCCACTCGA-G-----------------------------------------------------------------------------------------------------TTCC----G----AT--------GAGCCTTTTTC--------T---------GGAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGGGT-----TAAT------ACTTCCT-TGGATGTCTGATC-------------
Tree/1-101 ---------TTTGTGATGAAAG----TTTCCATT---CTATTCGCAGTTTCCACTCGA-G-----------------------------------------------------------------------------------------------------TTCC----G----AT--------GAGCCTTTTTC--------T---------GGAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGGGT-----TAAT------ACTTCCT-TGGATGTCTGATC-------------
Tatr/1-101 ---------TGCATGATGAAAT----TTTCCATT---TAATTCGCAGTTTCCACTCGA-G-----------------------------------------------------------------------------------------------------TTCC----G----AT--------GAGCCTTTTTC--------T---------GGAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGGGT-----TAAT------ACTTCCT-TGGATGTCTGATC-------------
Nhae/1-101 ---------TGCATGATGAAAT-----TTCCATAC--ACATTCGCAGTTTCCACTTGA-G-----------------------------------------------------------------------------------------------------TCCC----G----ATC-------GAGCCTTTTTT------------------GGGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AAATGAAGGT-----TTAT------TCTTCCT-TGGATGTCTGATC-------------
Foxy/1-101 ---------TGCATGATGAAAA----TTTCCTCT---TAATTCGCAGTTTCCACTCGA-G-----------------------------------------------------------------------------------------------------TCCC----G----ATC-------GAACCTTTTC---------T---------GGGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGGGT-----TTAT------TCTTCCT-TGGATGTCTGATC-------------
Gzea/1-104 ---------TTCATGATGAAAT----TTTCCACC---TAATTCGCAGTTTCCACTCGA-G-----------------------------------------------------------------------------------------------------TCCC----A----TTC-------GAGCCTTTTT-------------------GGGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGGGTAT---AATT----ATACTTCCT-TGGATGTCTGATT-------------
Vdah/1-100 ---------TCGGTGATGAAAG-----TTCCTACCTTAACTTCGCAGTTTCCTCTCGA-G-----------------------------------------------------------------------------------------------------CTTC---------T-------AAGAGCCTTTT--------------------GAAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AAATGAGGGT-----TTAC------ACTTCCT-TGGATGACTGAGA-------------
#=GS Fful/1-114 DE tomato leaf mould CP090163.1:c3301468-3301355 Fulvia fulva strain Race5_Kim chromosome 1
Fful/1-114 ----TCCGCTGGATGATGAAAA--ACTTTTCC-----ACATTCGCAGTTTCCACTCGA-G----------------------------------------------------------------------------------------------------CCATC----G----ACGC------GAACCTACTT---------A---------GATGG------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGGGG-----CAAT------TCTTCCT-TGGATGTCTGACCCAGCT--------
#=GS Bgra/1-97 DE LR026992.1 Blumeria graminis f. sp. tritici
Bgra/1-97 -----------CATGATGAAAA---ACATCA------CCATTCGCAGTTTTCACTCGA-G-----------------------------------------------------------------------------------------------------TCCC----A----ACTTA-----GAACCTATTTC------------------GGGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGGGC-----CAAT------TCTTCCT-TGGATGTCTGA---------------
#=GS Shom/1-105 DE JW824688.1 TSA: Sclerotinia homoeocarpa 11369.Schocontig
Shom/1-105 --------TTCGATGATGAAAC--ACATACC------ACATTCGCAGTTTTCACTCGA-G----------------------------------------------------------------------------------------------------TCCTC----G----CCT-----TAGAACCTAAACA------------------GAGGA------------------------------------------------------------------------------------------------------------------------------------------------------------------T----AAATGAGGGC-----CAAT------TCTTCCT-TGGATGTCTGATT-------------
Sscl/1-103 ---------TTCATGATGAAAC--ACATACC------ACATTCGCAGTTTTCACTCGA-G----------------------------------------------------------------------------------------------------TCCTC----G----CCT-----TAGAACCTAAATA------------------GAGGA------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGGGT-----CAA-------TCTTCCT-TGGATGTCTGATT-------------
Bfuc/1-103 ---------ACTATGATGAAAC--ACATACC------ACATTCGCAGTTTTCACTCGA-G----------------------------------------------------------------------------------------------------TCCTC----G----CCT-----TAGAACCTAAATA------------------GAGGA------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AGATGAGGGT-----ATA-------TCTTCCT-TGGATGTCTGATC-------------
#=GS Glgr/1-135 DE ET530419.1 fcg3x.226750c02 C. graminicola genomic sequence Glomerella graminicola
Glgr/1-135 ---------AACATGATGAAAT--ATCCACC------AAATTCGCAGTTTTCACTCGA-GTCAC--------------------------------------------G-----------------------------------------------------GCC---------AC--------GAGCTTTTC----------C---------GGC--------------------------------------------------------GGGCGCTTCTTT----TCA--AAAGAGGGCGTCT-----------------------------------------------TAA-----------------------GTGAT-GCATGAGGGC-----CAAT------TCTTCCT-TGGATGTCTGACC-------------
Lbic-b/1-216 ---------TTCATGATGATAA------ACG---ACCACATTCGCAGTTTCCACTCTA-GCGCC--CTCCG----TT----TGGAG-T----ACTCCCCCAA----TGCGGAACG-----TCTATATT---------------------CCA--C--CTCCTC----------------GTGCTGAACCCAAAAGGCAT-----T--------GAGGAGTTC-T-TGG----C-------------------------------------------CG--CCAT----TC----GTGG-TG-------------------------------------TGCGCTGGG-CGGGATGCGGGGG----TC--------------GGCCCGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGATCTTTTT--------
#=GS Lbic-a/1-221 DE LACBIscaffold_2 dna:supercontig supercontig:V1.0:LACBIscaffold_2:1488713:1489127:-1
Lbic-a/1-221 ----TCATCTCAATGATGATAA------ACG---ACCACATTCGCAGTTTCCACTCTA-GCGCC--CTCC-----TTT----GGAG-T----ACTCCCCCAA----TGCGGAACG-----TCTATATT---------------------CCA--C--CTCCTC----------------GTGCTGAACCCAAAAGGCAT-----T--------GAGGAGTTC-T-TGG----C-------------------------------------------CG--CCAT----TC----GTGG-TG-------------------------------------TGCGCTGGG-CGGGATGCGGGGG----TC--------------GGCCCGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGATCTTTTTT-------
#=GS Lcas/1-220 DE ENA|MK705798|MK705798.1 Lepiota castanea voucher RITF2403 25S ribosomal RNA gene and 25S-5S ribosomal RNA intergenic spacer, partial sequence.
Lcas/1-220 --------TTGCAAGATGAAAA-----TCTGGT-CCATCATTCGCAGTTTCCACTCTA-GCACC--CTCC----CTTCCT--GGAG-T----ACTCCCCCAA----TGTGGAACA----TCC-ATATT---------------------CCT--C--CTTGT------TT--------GTATGTGAACCCAATATGCATGC-------------ACAAG--TGT-AGG----C-------------------------------------------CA--CCAT---GCT----GTGG-TG---------------------------------------CGCTGGG-TGGGAAGCGGGGGG--TTT--------------GGTGTGATGCGGGGTC---AATA----GACCTTCCTT3GGATGTCTGATA-------------
#=GS Lven/1-219 DE RCFS01000037.1:400717-400935 Lepiota venenata strain LJE Contig37, whole genome shotgun sequence
Lven/1-219 --------TTGCATGATGAAAA--TCATAT----CCATCATTCGCAGTTTCCACTCTA-GCACC--CTCCT----TTC---GGGAG-T----ACTCCCCCAA----TGCATAACA----CTC-ACATT---------------------CCT--C---TT-------TTT----T---GCATGGGAACCCAAAACACATGC---TT----------AA--TTGT-AGG----C-------------------------------------------CA--CC----ACCTAC----GG-TG---------------------------------------CGCTGGG-TGGGGAGCGGGGG----TT--------------GGTGTGATGCGAGGTC---ATTA----GACCTTCCTTTGGATGTCTGATATGCAT--------
#=GS Pabs/1-230 DE ENA|AB287002|AB287002.1 Pholiota abstrusa genes for 28S rRNA, IGS1, 5S rRNA, partial and complete sequence, clone: ATCC48187_IGS1_061013_6.
Pabs/1-230 ----TGAATAAGGTGATGAAAA------ACCGT-ATATCATTCGCAGTTTCCACTCTA-GTGTC--CTCCT----TA----GGGAG-C----ACTCCCCCAA----CGCGGAACG----CTTACTATT---------------------CCC--C--CTCGCT-----CT---------GTGTTGAACCCAATTCTATCAC---TA-------GGCGAG--CGT-GGG----C-------------------------------------------CG--CCA---CTTTGT---TGG-TG---------------------------------------CGCTGAG-CGGAAAGCGGGGG----TC--------------GACACGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGACTCTCATTC------
#=GS Psil/1-224 DE ENA|HF912366|HF912366.1 Psilocybe silvatica genomic DNA containing partial 28S rRNA gene, 28S rRNA-18S rRNA IGS, strain P24, specimen voucher DAOM:187848
Psil/1-224 ---TTTGAACGGGTGATGAAAA------ACCG--TAATCATTCGCAGTTTCCACTCTA-GTGTC--CTCC-----TC-----GGAG-T----ACTCCCCCAG----TGCGGAACG----CTTCACATT---------------------CCC--C--CTTG---------------CT-GTGTTGAACCCAAAAGACATTAG-------------CAAGCCTGT-GGG----CC------------------------------------------GG--CCG---GTAACA---CGG-TT-------------------------------------TGCGCTGAG-CGGAAAGCGGGGG----TC--------------GACACGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGATCCTT----------
#=GS Paur/3-226 DE ENA|AB286998|AB286998.1 Pholiota aurivella genes for 28S rRNA, IGS1, 5S rRNA, partial and complete sequence, clone: ATCC42988_IGS1_061011_7.
Paur/3-226 ---TGAAATAGGGTGATGAAAA------ACCGT-GTATCATTCGCAGTTTCCACTCTA-GTGCC--CTCCGT---AAA---ACGAG-T----ACTCCCCCAA----TGCGGAATG----CTTTATATT---------------------CCC--C--CTTGA------TT--------AGTGTTGAACCCAAAAGATACT--------------TCAAG--CGT-GGG----C-------------------------------------------CG--CCA----TTA-----TGG-TG---------------------------------------CGCTGAG-CAGAAAGCGGGGG----TT--------------GGCACGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGACCTCTTT--------
#=GS Pnam/1-223 DE AB286993.1:613-835 Pholiota nameko genes for 28S rRNA, IGS1, 5S rRNA, partial and complete sequence, clone: kawamura_KN33_2_IGS1_061010_9
Pnam/1-223 --------TAGAGTGATGAAAA------ACCGT-GAATCATTCGCAGTTTCCACTCTA-GTGCC--CTTCTT---TG---AAGGAG-T----ACTCCTCCAA----TGCGGAATG----TCTTATAAT---------------------CCC--C--CTTGATT--------------AGTATTGAACCCATTTAATGCT----T-------AA-CAAG--TGT-GGG----C-------------------------------------------CA--CCA----TTA-----TGG-TG---------------------------------------CGCTGGG-CAGAAAGCGGGGG----TT--------------GGCACGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGATCTTTTTCA------
#=GS Pspu/1-221 DE AB287000.1:625-845 Pholiota spumosa genes for 28S rRNA, IGS1, 5S rRNA, partial and complete sequence, clone: Ph_spumosa_daisen_IGS1_061011_10
Pspu/1-221 --------TGGCGTGATGAAAA------ACCGT-GTATCATTCGCAGTTTCCACTCTA-GTGCC--CTCT----GAAA----GGAG-C----ACTCCTCCAA----TGTGGAACG----CTTTATATT---------------------CCC--C--CTTG--------------ACTAGTGTTGAACCCAAAAGGCACT----GTG--------CAAG--TGT-GGG----C-------------------------------------------CG--CCA-----TA-----TGG-TG---------------------------------------CGCTGAG-CGGAAAGCGGGGG----TT--------------GGCACGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGATCTCTAGTAA-----
#=GS Pgal/1-223 DE JAIFHD010004692.1:c568-346 Psilocybe galindoi strain Georgia-2-SW k141_13369, whole genome shotgun sequence; same as JAILYQ010002523.1:c3760-3540 Psilocybe tampanensis isolate sample1 k141_649, whole genome shotgun sequence
Pgal/1-223 -----TTTAAAAGTGATGAAAA-----GACC---TTTTCATTCGCAGTTTCCACTCTA-GTGTC--CTCT-----TC-----GGAG-T----ACTCCCCCAA----TGCGGAACG----CTTTACATT---------------------CCC--C--CTTG-------CT---------ATGTTGAACCCAAAAGACAT-----TAG--------CAAGCCTGT-GGG----C-------------------------------------------CGGACC-TG--TAA--CAGGGTCTG---------------------------------------CGCTGGG-CGGAAAGCGGGGG----TT--------------GACACGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGATCTTT----------
#=GS Gmar/1-215 DE AYUM01001090.1:738-952 Galerina marginata CBS 339.88 GALMAscaffold_102_Cont1090, whole genome shotgun sequence
Gmar/1-215 ----TGACACAAGTGATGAAAA--AAGGT-----GTATCATTCGCAGTTTCCACTCTA-GTGTC--CTCT-----TCT----AGAG-T----ACTCCCTCAA----TGCAGAACG----CTTTACATT---------------------CCC--C--CTTGC----------------TGTGTTGAACCCAAAAGACACA----A---------GTAAG-AGTT-GGG----C-------------------------------------------CG--CCA-----TA-----TGG-TG---------------------------------------CGCTGGG-CGGAAAGCGGGGG----TT--------------GACACGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGATCT------------
#=GS Pazu/1-227 DE JAIFHE010056956.1:c2851-2625 Psilocybe azurescens strain Oregon-Coast-3-SW k141_106247, whole genome shotgun sequence
Pazu/1-227 ---------TGAGTGATGAAAA------AACG--TATTCATTCGCAGTTTCCACTCTA-GTGTC--CTCC----ACAA----GGAG-T----ACTCCCCCAA----TGCCGAACG----CTTTACATT---------------------CCC--C--CTTGTGTG----------------TGTGAACCCAAGTGTA---ACAACTTGAT-CATGCAAG-CTGT-GGG----C-------------------------------------------CT--ACCG-CTAAACC--CGGT-TG---------------------------------------CGCTGAG-CGGAAAGCGGGGG----TC--------------GACACGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGACCCT-----------
#=GS Hcru/1-217 DE AF174457.1:c348-132 Hebeloma crustuliniforme icg 3 strain 686 25S/5S rRNA intergenic spacer region
Hcru/1-217 --------TTGCTTGATGAAAA------ACCG--TTCACATTCGCAGTTTCCACTCTA-GTGTC--CTC--A--TTTTA----GAG-C----ACTCC-CCAG----TGCGGAACA----CCTCATAAT---------------------CCC--C--CTTGT------T----------GTGTTGAACCCAAAAGACAT-----TA--------GCAAG--TGT-GGG----C-------------------------------------------CG--CC----TTCACC----GG-TG---------------------------------------CGCTGGG-TGGAAAGCAGGGG----TT--------------GACACGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGACCCACTTGG------
#=GS Hhel/1-221 DE ENA|AF174458|AF174458.1 Hebeloma helodes icg 20 strain 688 25S/5S rRNA intergenic spacer region.
Hhel/1-221 ----TGAATTGCTTGATGAAAA------ACCG--TTCACATTCGCAGTTTCCACTCTA-GTGTC--CTC--A--TTTTA----GAG-C----ACTCC-CCAG----TGCAGAACA----CCTCATAAT---------------------CCC--C--CTTGT------T----------GTGTTGAACCCAAAAGACAT-----TA--------GCAAG--TGT-GGG----C-------------------------------------------CG--CC----TTCACA----GG-TG---------------------------------------CGCTGGG-TGGAAAGCAGGGG----TT--------------GACACGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGACCCACTTGG------
#=GS Pgig/1-216 DE AZAG01000138.1:21923-22138 Phlebiopsis gigantea 11061_1 CR5-6 PHLGIscaffold_13_Cont138, whole genome shotgun sequence
Pgig/1-216 ------TTTTGGATGATGAAAC----ATTC----CGTACATTCGCAGTTTTCACCCTA-GCGCC--CTCCT---TAAC---GGGAG-C----ACTCCTCCAA----TGCGGAACG----TC-TACATT---------------------CCC--C--CCTCA----------------TGTGCTGAACCCAAAAGGCATG----T---------TGAGG--TGT-GGG----C-------------------------------------------CG--CCC-----TTC----GGG-TG---------------------------------------CGCTGGG-CGGAAAGCGGGGG----TC----------------GGCGCGGTGAGGGC--CACAA---GTACCTTCCTTTGGATGTCTGACCCTT----------
#=GS Badu/1-213 DE BSCZ01002279.1:c457767-457555 Bjerkandera adusta Dec 1 DNA, scaffold2279_cov14, whole genome shotgun sequence
Badu/1-213 ---------TCTATGATGAGTA------T-----TCCACATTCGCAGTTTTCACTCTA-GCGCC--CTCTC---GAAAA--GGGAG-T----ACTCCCCCAA----TGCGGAACG-----TCTATATT---------------------CCC--C--CTTC-------T-------GCTGTGCGGAACCCAATATCGTGCAGC------------GAAG--TGT-GGG----C-------------------------------------------CG--CCC----AAT-----GGG-TG---------------------------------------CGCTGGG-CGGAAAGCGGGGG----TC----------------GGCGCGGTGAGGGT--CCGAA---GTACCTTCCTTTGGATGTCTGACTCT-----------
#=GS Cput/1-219 DE XM_007767362.1:368-586 Coniophora puteana RWD-64-598 SS2 hypothetical protein mRNA
Cput/1-219 --------TTTTATGATGAAAC---TTTTC----CGTACATTCGCAGTTTCCACTCTA-GAGCC--CTCCT---CGCA--AGGGAG-T----ACTCCCCCAA----TGCGGAACG----TC-TACATT---------------------CCC--C--CGCGCCC-----------------CGTGAACCCAAAAACG-------T-------GGGCGTG--TGT-GGG----C-------------------------------------------CG--CCCC----TC----GGGG-TG---------------------------------------CGCTGGG-CGGGAAGCGGGGG----CT--------------GGCTCGATGAGAGGTCT--TCGC---AGACCTTCCTTTGGATGTCTGATCCTTCT--------
#=GS Gnec/87-318 DE XM_043180902.1:1-321 Guyanagaster necrorhizus MCA 3950 uncharacterized protein (BT62DRAFT_44257), mRNA
Gnec/87-318 ---TCCACCCACCGGATGAAAA----TTTCC---GTTACATTCGCAGTTTCCACTCTA-GTTGCC-CTCT----TTAT----GGAG-T----ACTCCCCCAA----TGCGGAACG----TC-TATATT---------------------CCC--C--CTCCATA----C---------CAACCTGAACCCATTGGGTTG-------------TGTCGAG--CGT-GGG----C-------------------------------------------CG--CCCC---TTTG---GGGG-TG---------------------------------------CGCTGGG-CGGGAAGCGGGGG----TC--------------GGCACGATGCGGGATG---ATAA---TGTCCTTCCTTTGGATGTCTGATCCCTCTCCTC----
#=GS Palb/1-212 DE JAMZQW010013150.1:4845-5056 Pisolithus albus isolate MSR1 232204, whole genome shotgun sequence
Palb/1-212 ---------TTCACGATGAAGT--ATATTC----CGTTTATTCGCAGTTTCCACTCTA-GTGCC--CTCCA----CG----TGGAG-T----ACTCCCCCAA----TGCCGAACG-----TTTATATT---------------------CCC--C--CTTG-------T---------GCTGTGGAACCCATTAACATGGC--------------CAAG-A-GT-GGG----C-------------------------------------------CG--CCT-----TC-----GGG-TG---------------------------------------CGCTGAA-CGGGAGGCGGGGG----TC--------------GGCGCAATGAGGGAC---CACAAA--GTTCCTTCCTTTGGATGTCTGATCCT-----------
#=GS Aost/1-217 DE LR732075.1:4579013-4579229 Armillaria ostoyae strain C18/9 genome assembly, chromosome: LG1
Aost/1-217 ----TCTGTCGCACGATGAAAA---ATTTC----CGTACATTCGCAGTTTCCACTCTA-GCGCC--CTT------TT------GAG-T----ACTCCCCCAA----TGTGAAACG----CT-TATATT---------------------CCC--C--CTCCAT-----AC--------CATGTTGAACCCATTGACATG-----C--------GTCGAG--TGT-GGG----C-------------------------------------------CG--CCC----TTT-----GGG-TG---------------------------------------CGCTGG--CGCGGATCGGGGG----TC--------------GGCGCGATGCGAGTTG----CAA---TGACCTTCCTTTGGATGTCTGATTCTTCT--------
#=GS Gand/1-213 DE VKGB01000399.1:c27987-27775 Gymnopus androsaceus JB14 BT96scaffold_142_Cont399, whole genome shotgun sequence
Gand/1-213 -------TATACAAGATGAAAT----TTTC----CGCACATTCGCAGTTTCCACTCGA-GCGTC--CA-------TTT-----TAG-T----ACTCCCCCAA----TGCGGAACG-----TTTATATT---------------------CCC--C--CTCTAT-----AC---------CATGTGAACCCAAAAGCATG-----C----------GTTGAGTGT-GGG----C-------------------------------------------CG--CTTC---TTCG---GAGG-TG---------------------------------------CGCTGG--CGGGAAGCGGGGG----TC--------------GGCGCCATGAGGGTTG----TAA---TGTCCTTCCTTTGGATGTCTGAACTTT----------
#=GS Ldec/1-214 DE CP107713.1:c214778-214565 Lyophyllum decastes strain LRG-d1-1 chromosome 13
Ldec/1-214 ------ACATGCTTGATGAAAC------ACA---CCTACATTCGCAGTTTCCACTCTA-GCGTC--CTCT-----TC-----GGAG-C----ACTCCCCCAA----TACCGAACA----T-TTATTCT---------------------CCC-CCT-TGGTT-----------------GTGTTGAACCCAAAAGACAC----ACCG-------AGCCG--C---GGG----C-------------------------------------------CG--CCC---TTTTT----GGG-TG---------------------------------------CGATGA--TGCAAGGTGGGGG----TC--------------GACGCGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGACCCATT---------
#=GS Ter/1-207 DE KQ412244 Termitomyces sp. J132 supercontig:TerJ1321.0:KQ412244:33740:33953:-1
Ter/1-207 ---------TTCTTGATGAAAC--GCTT------CCTACATTCGCAGTTTCCACTCTA-GCGTC--CTCC-----TTT----GGAG-T----ACTCCCCCAA----TACCGAACA----TTCATTCTC---------------------CCC-CT--CTTTG-------------------GTCGAACCCAAAGATC-------A---------CAGAG-TTGT-GGG----C-------------------------------------------CG--CC----CACGT-----GG-TG---------------------------------------CGATGA--TGCTAGGTGGGGG----TC--------------GACGCGATGCGAGTTG----AAA---CGACCTTCCTTTGGATGTCTGACCCTTT---------
#=GS Tful/1-217 DE AB619792.1:680-896 Tricholoma fulvocastaneum DNA, Basidiomycota specific DNA element megB1, complete sequence, strain: Tf-AM-7
Tful/1-217 ---------TCTATGATGAAAT----ATAC----CATACATTCGCAGTTTCCACTCTA-GTGTC--CTCCA---GCAA---TGGAG-C----ACTCCTCCAG----TACCGAACA----TC-CATATT---------------------CCC--C--CTCATG-----CG----------TGTCGAACCCAAAAGACA------A--------CATGGG-CTGT-GGG----C-------------------------------------------CG--CC----CTTTGT----GG-TG---------------------------------------CGCTAG-GTGCGAGGTGGAGG----TC--------------GACACGATGCGGGCCG----CAA---TGACCTTCCTTTGGATGTCTGATCCTTCT--------
#=GS Tmak/1-218 DE AB304914.1:2479-2696 Tricholoma matsutake DNA, macroevolutionary genomic marker from Basidiomycota megB1, tRNA-Pro like sequence, complete sequence, strain: Y1 (= IFO 33136)
Tmak/1-218 ---------TTTATGATGAAAC----ATAC----CATACATTCGCAGTTTCCACCTTA-GTGTC--CTCCA---GCGA---TGGAG-C----ACTCCTCCAA----TACCAAACA----TC-CATATT---------------------CCC--C--CTCATG-----C----------GTGTTGAACCCAAAAGACAC--------------CATGAG-CTGT-GGG----C-------------------------------------------CG--CC----CTTTGT----GG-TG---------------------------------------CGCTAG-GTGCGAGGTCGGGGG---TC--------------GACACGATGCGGGTCG----CAA---TGACCTTCCTTTGGATGTCTGATCCTTCT--------
#=GS Tana/1-223 DE AB543748.1:450-672 Tricholoma anatolicum DNA, Basidiomycota specific DNA element: megB1, clone: pCR2.1::2A-Out-megB1-TaS231
Tana/1-223 ----TCCAATTTATGATGAAAC----ATAC----CATACATTCGCAGTTTCCACCTTA-GTGTC--CTTCA---GAAA---TGGAG-C----ACTCCTCCAA----TACCAAACA----TC-CATATT---------------------CCC--C--CTCATG-----C----------GTGTTGAACCCAAAAGACAC--------------CATGAG-TTGT-GGG----C-------------------------------------------CG--CT----CTTTGT----GG-TG---------------------------------------CGCTAG-GTGCGAGGTCGGGGG---TC--------------GACACGATGCAGGTCG----CAA---TGACCTTCCTTTGGATGTCTGATCCTTCT--------
#=GS Iflo/1-229 DE OM964556.1:2280-2508 Inocybe flocculosa voucher EL168-16 small subunit ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and large subunit ribosomal RNA gene, partial sequence
Iflo/1-229 ------TTGAGCATGATGAAAA--GCTTGCC---TATGTATTCGCAGTTTCCACTCTA-GCACCG-TGTT------------GAGA------ACTCCCCCAG----TGCAGAACG----CTTTACATT---------------------CCC--C--CTTGATCTTG--------------CTTGAACCCCAAG---------GAA---CAAGA-CAGG--TGTTGGG----C-------------------------------------------CG--CCAA---TGAT---TTGG-TG---------------------------------------CGCTGA-GCGGAAAGCGGGGGT---------------TGGTGCTGTGAGAGATCATTG---AAA---CGACCTTCCTTTGGATGTCTGAGCTCAAGGCA-----
#=GS JEC21_CND00700/67-335 DE chromosome:ASM9104v1:4:195170:195794:1
JEC21_CND00700/67-335 ------TTTTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCCCTTTTCTCCTCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TT--------CCATGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACA----GATTC----CTCGACCTCGG--GCTC------------------------------------GTAA------------------------------------------GAGC--AGTCGGTACGATCG--------TTGGGG----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TTTCTTCCT-TGGATGTCTGATTCCTTTTTTT----
#=GS XL280alpha/1-268 DE CP072814.1:194715-194982 Cryptococcus neoformans var. neoformans strain XL280alpha chromosome 04
XL280alpha/1-268 ------TTTTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCACTTTTCTCCTCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TT--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACA----GATTC----CTCGACCTCGG--GCTC------------------------------------GTAA------------------------------------------GAGC--AGTCGGTACGATCG--------TTGGGG----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TTTCTTCCT-TGGATGTCTGATTCCTTTTTT-----
#=GS H99_CNAG_00930/61-329 DE chromosome:CNA3:5:1603206:1603823:-1
H99_CNAG_00930/61-329 ------TTTTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCCCTTCTTTCGTCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TG--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACA----GATTC----CTCGACCTCGG--GCTC------------------------------------GCAA------------------------------------------GAGC--AGTCGGTGGGATCG--------TTGAGG----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TCTCTTCCT-TGGATGTCTGACTCCTATTTTA----
#=GS Bt65/1-269 DE CP097914.1:c1686576-1686308 Cryptococcus neoformans strain Bt65 chromosome 5
Bt65/1-269 ------TTTTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCCCTTCTCTCGTCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TG--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACA----GATTC----CTCGACCTCGG--GCTC------------------------------------GCAA------------------------------------------GAGC--AGTCGGTGGGATCG--------TTGAGG----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TCTCTTCCT-TGGATGTCTGATTCCTATTTTA----
#=GS VNII/1-269 DE CP091249.1:c1664756-1664488 Cryptococcus neoformans strain VNII chromosome 3
VNII/1-269 ------TTTTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTTCTCGCCCTTCTCTCGTCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TG--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACA----GATTC----CTCGACCTCAG--GCTC------------------------------------GCAA------------------------------------------GAGC--AGTCGGTGGGATCG--------TTGAGG----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TCTCTTCCT-TGGATGTCTGATTCCTATTTTA----
#=GS EN28/1-269 DE CP025721.1:c1592505-1592237 Cryptococcus neoformans strain EN28 chromosome 5
EN28/1-269 ------TTTTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCCCTTCTCTCGTCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TG--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACA----GATTC----CTCGACCTCAG--GCTC------------------------------------GCAA------------------------------------------GAGC--AGTCGATGGGATCG--------TTGAGG----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TCTCTTCCT-TGGATGTCTGATACCTATTTTA----
#=GS IND107/67-335 DE Cryp_gatt_IND107_V2:supercont2.24:170567:171191:1
IND107/67-335 ------TACTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCCCTTCTCACATCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TT--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACG----GATTC----CTCGACCTCGG--GCTC------------------------------------GCAA------------------------------------------GAGC--AGTCGGTGGGATCG--------TTGGGG----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TTTCTTCCT-TGGATGTCTGATTCCTTTTTTT----
#=GS WM276/1-269 DE CP000295.1:c322341-322073 Cryptococcus gattii WM276 chromosome J
WM276/1-269 ------CACTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCCCTTCTCACATCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TT--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACG----GATTC----CTCGACCTCGG--GCTC------------------------------------GCGA------------------------------------------GAGC--AGTCGGTGGGATCG--------TTGGGG----AGGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TTTCTTCCT-TGGATGTCTGACTCCTTCTTTT----
#=GS EJB2/67-335 DE Cryp_gatt_EJB2_V1:supercont1.105:9492:10115:-1
EJB2/67-335 ------CACTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCCCTTCTCACATCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TT--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACG----GATTC----CTCGACCTCGG--GCTC------------------------------------GCGA------------------------------------------GAGC--AGTCGGTGGGATCG--------TTGGGG----AGGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TTTCTTCCT-TGGATGTCTGACTCCTTCTTTT----
#=GS CA1280/67-335 DE Cryp_gatt_CA1280_V1:supercont1.23:174125:174748:1
CA1280/67-335 ------CACTTTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCCCTTCTCACATCGGTCTCGCTT-CTGAAA-CCGAG-TT---GAGTC------TT--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACG----GATTC----CTCGACCTCGG--GCTC------------------------------------GCGA------------------------------------------GAGC--AGTCGGTGGGATCG--------TTGGGG----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TTTCTTCCT-TGGATGTCTGACTCCCTTCTTT----
#=GS CryVGI/1-262 DE JACUVQ010000138.1:c24686-24425 Cryptococcus gattii VGI strain WM1802 sc33, whole genome shotgun sequence
CryVGI/1-262 ---------TCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGTCCTTTTCCACATCGGTCTCGCTTCTGAAA-CTGAG-TT---GAGTC------TT--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACG----GATTC----CTCGACCTCGG--GCTC------------------------------------GCGA------------------------------------------GAGC--AGTCGGTGGGATCG--------TTGGGA----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TTTCTTCCT-TGGATGTCTGACTCCTT---------
#=GS R265/1-269 DE CP025772.1:174306-174574 Cryptococcus gattii VGII R265 chromosome 14
R265/1-269 ------CACTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCCTTTCTCATTTCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TT--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACC----GATTC----CTCGACCTTGG--GCTT------------------------------------GCAA------------------------------------------GAGC--AGTCGGTGAGATCG--------TTGGGG----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TTTCTTCCT-TGGATGTCTGATTCCTTTCTCT----
#=GS Ram5/67-335 DE Cryp_gatt_Ram5_V1:supercont1.19:344394:345018:-1
Ram5/67-335 ------CACTCTATGATGAAACCCA-ATGT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----TGCC--GGGAG-TTTGTTCCTCGCCTTTCTCATTTCGGTCTCGCTT-CTGAAA-CTGAG-TT---GAGTC------TT--------CCTTGGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCTTTGG-----ACC----GATTC----CTCGACCTTGG--GCTT------------------------------------GCAA------------------------------------------GAGC--AGTCGGTGAGATCG--------TTGGGG----AAGGGCTT-GGC---GATGC--TAG-TTTGAAGTTT-TTTCTTCCT-TGGATGTCTGATTCCTTTCTCT----
#=GS CryVH57/1-259 DE JANBXI010000298.1:1-259 Cryptococcus sp. VH57_BW 299, whole genome shotgun sequence
CryVH57/1-259 ---------------ATGAAATGTT-TTATC--AAAACCATACGCAGTTTCCACTGATCGCGTC--CTCCT----TGCC--GGGAG-TTTGT-GCTTGCTATCTCTTGTCTGCTTCCGCTT-CCGTAA-CTGAG-TC---GAGTC------TT--------CTTCTGTTC--------CGGGTTGAACCTCCCG-----TT---GAACATTGG-----ATA----GATTC----CTCGATCTCGG--GCTC------------------------------------GCAA------------------------------------------GAGC--AGTCGGCGGAAC-G--------TAGACG----GAGGAGCGAGCC---GATGC--TAA-CTTGAAGTTT-TTTCTTCCT-TGGATGTCTGATCCCCCCCCT-----
#=GS CryVH57/1-260 DE JANBXI010000081.1:65360-65619 Cryptococcus sp. VH57_BW 81, whole genome shotgun sequence
CryVH57/1-260 --------TTTTATGATGAAATGTT-TTATC--AAAACCATACGCAGTTTCCACTGATCGCGTC--CTCCT----TGCC--GGGAG-TTTGT-GCTTGCTATCTCTTGTCTGCTTCCGCTT-CCGTAA-CTGAG-TC---GAGTC------TT--------CTTCTGTT--------ACGGGCTGAACCTCCCGT----TG----AACATTGG-----ATA----GATTC----CTCGATCTCGG--GCTC------------------------------------GCAA------------------------------------------GAGC--AGTCGGCGGAAC-G--------TAGACG----GAGGAGCGAGCC---GATGC--TAA-CTTGAAGTTT-TTTCTTCCT-TGATTGTCTGATCCC-----------
#=GS Camy-2/1-269 DE XM_019136338.1:100-367 Cryptococcus amylolentus CBS 6039 argininosuccinate synthase, variant 2 (L202_02683), mRNA
Camy-2/1-269 ------CTTTTCATGATGAGAACCA--TTTC--AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----CGCC--GGGAG-TTTGT-CCTCGTCCTTTC--TTTCGGCACTGTCC-CTGTAA-CCGAG-TC---GAGTC------GA--------CTCTAGTTC--------CAAGTAGAACCTCTTG-----TT---GAGCTGGAG-----AC-----GATTC----CTCAATCTCAG--GCTTGGA---------------------------------GCAA---------------------------------------TCCGAGC--AGCCGACGCTGTCG--------ACAAGG----AAGGACTT-GGC---GATGC--TAG-TTTGAAGTTT--CACTTCCT-TGGATGTCTGAACCCATTTTA-----
#=GS CryAmy/59-328 DE Cryp_amyl_CBS6273_V2:supercont2.4:219039:219655:-1
CryAmy/59-328 ------CTTTTCATGATGAGAACCA--TTTC--AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----CGCC--GGGAG-TTTGT-CCTCGTCCTTTC--TTTCGGCACTGTCC-CTGTAA-CCGAG-TC---GAGTC------GA--------CTCTAGTTC--------CAAGTAGAACCTCTTG-----TT---GAGCTGGAG-----AC-----GATTC----CTCAATCTCAG--GCTTGGA---------------------------------GCAA---------------------------------------TCCGAGC--AGCCGACGCTGTCG--------ACAAGG----AAGGACTT-GGC---GATGC--TAG-TTTGAAGTTT--CACTTCCT-TGGATGTCTGAACCCATTTTAT----
#=GS CryWin/1-270 DE CP034264.1:1386593-1386862 Cryptococcus wingfieldii strain CBS7118 chromosome 4
CryWin/1-270 ------TTTTTCATGATGAGAACCA--TTTC--AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----CGCC--GGGAG-TTTGT-CCTCGTTTTTTC--TTTCGGCACTGTCC-CTGTAA-CCGAG-TC---GAGTC------GA--------CTCTAGTTC--------CAAGTAGAACCTCTTG-----TT---GAGCTGGAG-----GC-----GATTC----CTCAATCACAG--GCTTGGA---------------------------------GCAA---------------------------------------TCCGAGC--AGCCGACGCTGTCG--------ACAAGG----AAGGACTT-GGC---GATGC--TAG-TTTGAAGTTT--CACTTCCT-TGGATGTCTGAACCCATTTTAT----
#=GS TsuWin/59-329 DE Tsuc_wing_CBS7118_V1:supercont1.35:96932:97548:-1
TsuWin/59-329 ------TTTTTCATGATGAGAACCA--TTTC--AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT----CGCC--GGGAG-TTTGT-CCTCGTTTTTTC--TTTCGGCACTGTCC-CTGTAA-CCGAG-TC---GAGTC------GA--------CTCTAGTTC--------CAAGTAGAACCTCTTG-----TT---GAGCTGGAG-----GC-----GATTC----CTCAATCACAG--GCTTGGA---------------------------------GCAA---------------------------------------TCCGAGC--AGCCGACGCTGTCG--------ACAAGG----AAGGACTT-GGC---GATGC--TAAGTTTGAAGTTT--CACTTCCT-TGGATGTCTGAACCCATTTTAT----
#=GS DioAur/1-250 DE BCKN01000002.1:467197-467446 Dioszegia aurantiaca DNA, scaffold: scaffold_1, strain: JCM 2956, whole genome shotgun sequence
DioAur/1-250 -------TTTCTATGATGATATACC--ATAC--AAAACCATACGCAGTTTCCACTGAA-GCGTC--CTCCT--TTCGCC--GGGAG-TTTGT-CCCTGTTT-CTTTCGGATCCCAGACCT-----TCATTCGGA-TC---GAGTC---------------ATGTCAGTTC---------CGGGTGAACCTCCTG-----TT---GAGCTGGCGT-----------GATTC----TCCGAGCGG----CCTC------------------------------------TCCG------------------------------------------GAGG--AGCCGGCT-TGTG--------GAGAAG----GGGAGACGG-GGC---GATGC--ATA-CTTGAAGTTT--TTCTTCCT-TGGATGTCTGACTCTT----------
#=GS ApiBra/1-245 DE JAMALJ010000038.1:c151757-151513 Apiotrichum brassicae strain M2204 ctg_38, whole genome shotgun sequence
ApiBra/1-245 ------TCTTCTATGATGAAATGTT--TTTT--AAAACCATACGCAGTTTCCACTGCA-GCGTC--CTCC----CTGCCA--GGAG-TTTGTT-CCCGCGCGTACAGCCCGCC-GAGCCT-----CTGCGGCGG-TC---GAGTGC----------------CCCGGTTC---------CGGTTGAACCTCCG------TT---GAGCCGGG-------C----GCATTC----CCCCGTGG-----GCTT------------------------------------TC--------------------------------------------GAGC--AGCCGGCAGACG---------CGGTGC----GGTCGCGTG-GGC---GATGC--T-A---TGAAGTTT-TTTCTTCCT-TGGATGTCTGACTCCCCTT-------
#=GS VanPse/1-259 DE CP086717.1:c2552374-2552116 Vanrija pseudolonga isolate DUCC4014 chromosome 4
VanPse/1-259 ------CTCTCTATGATGAAAACGT--TTTC--AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT--TTCGCC--GGGAG-TTTGT--CTCGCCAATCGCGCCGTT--GAGCCT-------CTGCGGG-TT---GAGTG----------------TCTCAGTTC--------CGGGTTGAACCTCCTG-----TT---GAGCTGTGG------G-----CATTC----CCTGCGG------GCGCCC----------------------------------GAAA----------------------------------------GGGTGC--AGCGTGGCAAGAC--------GTGCTG----TGGTCGGTG-AGC---GATGC--TTAATTTGAAGTCT-CTTCTTCCT-TGGATGTCTGATTCGCCCCCAT----
#=GS VanHum/1-249 DE BFAH01000028.1:c1419200-1418952 Vanrija humicola UJ1 DNA, scaffold29, whole genome shotgun sequence
VanHum/1-249 -------TCTCTATGATGAAAACGA--TTTC--AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT--TTCGCC--GGGAG-TTTGT--CTCGCCGATCGCGCCGCC--GAGCTT-------CTGCGGG-TT---GAGTGTC----T------------CAGTTC--------CGGGTTGAACCTCCTG-----TT---GAGCTG--------TG---GACATTC----CTCGCGG------GCGCCC----------------------------------GCAA----------------------------------------GGGTGC--AGCGTGGCAAGGT--------GTGCTG----TGGCCGGTG-AGC---GATGC--TCAATTTGAAGTCT-CTTCTTCCT-TGGATGTCTGATT-------------
#=GS TriAsa/81-333 DE Trichosporon_asahii_1:contig0194:129093:129715:1
TriAsa/81-333 ------CTCTCAATGATGATAATTT--CACCA-AAATCCATACGCAGTTTCCACTGA-CGCGTC--CTCCT--TTCGCC--GGGAG--TTGC---GCCGGTCGGGCGACGCCTCGTCTCT-------CTATGAG-TT---GAGTG------CA---------CTTGGCTC-------CGGATTTGAACCTTCTG-----TT---GAACCAAG-----AACG----CATTC----CTCATGG------GCCT-------------------------------------TC-------------------------------------------GGGC--AGCCGAGACGGG---------GCTGAG----CTCGTGCCG-GCT---GATGC---AAA---GAAGTCTTATTCTTCCT-TGGATGTCTGATTCATTCTCTA----
#=GS TriPB0/1-244 DE Jri0AMALZ010000019.1:507004-507247 Trichosporon sp. PB005A ctg_19, whole genome shotgun sequence
TriPB0/1-244 ------TCTTCAATGATGATAATTT--CACCA-AAATCCATACGCAGTTTCCACTGA-CGCGTC--CTCCT--TTCGCC--GGGAG--TTGC---GCCGGCCGGGCGACGCCCCGCCTCT-------CTGTGGG-TT---GAGTG------CA---------CTGGGCTC-------CGGACTTGAACCTTCTG-----TT---GAACCCAG-----AACG----CATTC----CCCACGG------GCCT-------------------------------------TC-------------------------------------------GGGC--AGCCGAGACGGA---------GCTGAG----CCCGCGCCG-GCT---GATGC---AAA---GAAGTCTTATTCTTCCT-TGGATGTCTGATT-------------
#=GS TriOvo/1-249 DE WEIQ01000010.1:1143304-1143552 Trichosporon ovoides strain 2NF903A scaffold10-size1595759, whole genome shotgun sequence
TriOvo/1-249 ----TTCTTTTAATGATGATATTAC---ACCA-AAATCCATACGCAGTTTCCACTGA-CGCGTC--CTCCT--TTCGCC--GGGAG--TTGC---GCTGTTCAGGCGAGGCTCTGTCTCT-------CTGTGGG-TT---GAGTG------CA---------CTTGGCTC-------CGGACTTGAACCTTCTG-----TT---GAACCAAG-----AACG----CATTC----CTCATGG------GCCT-------------------------------------TC-------------------------------------------GGGC--AGCCGAGATTGA---------GTTGAG----CCTGGGC-G-GCT---GATGC---AAA---GAAGTCTTATTCTTCCT-TGGATGTCTGATTCTTTT--------
#=GS CutCut/1-231 DE JAMALK010000062.1:24468-24698 Cutaneotrichosporon cutaneum strain P1411 ctg_62, whole genome shotgun sequence
CutCut/1-231 -----------TATGATGAACCAAC---ACC--AAAACCATACGCAGTTTCCACTGTA-GCGTC--CTCC----CTGCCA--GGAG-TAGT-T-CGTCGTCGTGCGCCGCGCT--------------CTGCGGG-TC---GAGTGC----------------CCCAGCC--------GCGGATTGAACCTTCTTGC----T----GGCTGGG-------C----GCATTC----CCTGTGG------GCCC------------------------------------GAAA------------------------------------------GGGC--AGCCGGCAACGCG--------TGCGGC----GGCGG-----CGA---GATGC--T----GTGAAGTTTTTTTCTTCCT-TGGATGTCTGATCTCTTCT-------
#=GS VanFra/1-252 DE BEDY01000003.1:c403100-402849 Vanrija fragicola DNA, scaffold: scaffold_3, strain: JCM 1530, whole genome shotgun sequence
VanFra/1-252 -------TCTCTCTGATGAAATTTA--TTTC--AAAACCATACGCAGTTTCCACTGATCGCGTC--CTCCT----TGCC--GGGAG-TTTGT-ACCTTTGCCGAGGTCGCAATTTGTCT--------TCACTGG-TC---GAGTGC-----C----------CTCGACTC--------CGGATTGAACCTTCTG-----TT---GAGTCGGG------AC----GCATTC----CCTTTGA------GCCTCGTC--------------------------------GCAA--------------------------------------GACGTTGC--AGCAGGCGGAGC---------GTGGCC----GAGGTATCG-GGC---GATGC--T-AGCTTGAAGTTT--TTCTTCCT-TGGATGTCTGATT-------------
#=GS CutCur/1-231 DE NIUX01000003.1:c898978-898748 Cutaneotrichosporon curvatum strain ATCC 10567 Contig003, whole genome shotgun sequence
CutCur/1-231 --------TTTCATGATGAATGTTT--TTC---AAAACCATACGCAGTTTCCACTGA-TGCGTC--CTCCG----CGC---CGGAG-TTGTC-CCTTCGGCCCAGGCTGCGCCT-------------CCAATGG-TC---GAGTGC----------------CCCGGCTC-------CGGATTTGAACCTTCTG-----TC---GAGTCGGG-------C----GCATTC----CCTGCGG------GC-------------------------------------TACCAT-------------------------------------------GC--AGACGGCAAGG----------CTGGCG----GTCGTTGGG-CGA---GATGC---AAA---GAAGCTT-TTTCTTCCT-TGGATGTCTGATTCT-----------
#=GS CutDer/1-235 DE JAMALL010000011.1:328059-328293 Cutaneotrichosporon dermatis strain M4303A ctg_11, whole genome shotgun sequence
CutDer/1-235 --------TCTCCTGATGAATTCCA--TTT---AAAACCATACGCAGTTTCCACTGTA-GCGTC--CTCC----CTGCCA--GGAG-TAGACTGCTAG-ATGTGCGCCGCGCT--------------CTGCGAG-TC---GAGTGC----------------CCCAGTTC--------CGGATTGAACCTTCTG-----TT---GAGCTGGG-------C----GCATTC----CTTGCGG------GCCCTC-----------------------------------TT-----------------------------------------GAGGGC--AGCCGGCAACGC---------GCATGT----CGACGCGGG-CGT---GATGC--T----TTGAAGTTT-TTTCTTCCT-TGGATGTCTGATTCT-----------
#=GS TriPL3/1-234 DE JAMAMB010000001.1:c3864084-3863851 Trichosporonaceae sp. PL3408A ctg_1, whole genome shotgun sequence
TriPL3/1-234 --------TCTCCTGATGAATTTTT--CTT---AAAACCATACGCAGTTTCCACTGTA-GCGTC--CTCC----CAGCCA--GGAG-TAGTCT-GCTCGGTGTGTACCGCGCCT-------------CTGTGGG-TC---GAGTGC----------------CCCGGTTC--------CGGATTGAACCTTCTG-----TT---GAGCCGGG-------C----GCATTC----CCCTCAG------GCCC-------------------------------------TC-------------------------------------------GGGC--AGCCGGCAACGT---------GCATGT----CGACGTGGG-CGA---GATGC--T----GTGAAGGTT-TTTCTTCCT-TGGATGTCTGATCCCTT---------
#=GS PasPL2/1-246 DE JAMFRE010000005.1:392256-392501 Pascua sp. PL2904B ctg_5, whole genome shotgun sequence
PasPL2/1-246 TACTCACCTAACCGGATGAAACTAT---GTTT-AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCC----CTGCCA--GGAG--TTG-A--CTGCCGACTCCCCGCCGCGCCT----------CTGCGAG-TC---GAGTGC----------------CCCAGGCG-------ACGGATTGAACCTCCTGT---------TGTCTGGG-------C----GCATTC----CTCGTGG------GCCCTCT----------------------------------TC----------------------------------------GGAGCGC--AGCCGGCAACGCG--------GGTTGA----GAAAGCGGG-CGC---GACGC--T-AA---GAAGTTT--TTCTTCCT-TGGATGTCTGATT-------------
#=GS TriInk/1-244 DE JXYM01000003.1:1826232-1826475 Trichosporon inkin strain JCM 9195 scaffold_0003, whole genome shotgun sequence
TriInk/1-244 -------TTTCAATGATGATACCAC---ACCA-AAATCCATACGCAGTTTCCACTGA-CGCGTC--CTCCT--TTCGCC--GGGAG--TTGT--GCCGTTCAGGTGAAGCTTCGTCCCT--------CTGTGGG-TT---GAGTG------CA---------CTTGGCTC-------CGGACTTGAACCTTCTG-----TT---GAACCAAG-----AACG----CATTC----CCCATAG------GCCT-------------------------------------TC-------------------------------------------GGGC--AGCCGGGACGGA---------GTCGAG----TCTGGGC-G-GCT---GATGC--C-AA---GAAGCCCTATTCTTCCT-TGGATGTCTGATCCTT----------
#=GS TriGue/1-253 DE BCJX01000006.1:c1105377-1105125 Trichosporon guehoae DNA, scaffold: scaffold_5, strain: JCM 10690, whole genome shotgun sequence
TriGue/1-253 -TCAAACCTCACCGGATGAAACCAT---GTTT-AAAACCATACGCAGTTTCCACTGAATGCGTC--CTCC----CTGCCA--GGAG--TTG-A-CTGCCGACTCCCCGCCGCGTCT-----------CTGCGAG-TC---GAGTGC-----------------CCCAGGC---TC---CAGATTGAACCTTCTG-----TT---GTCTGGG--------C----GCATTC----CTCATGG------GCCGCTCG--------------------------------TTCG--------------------------------------CGAGTGGC--AGCCGGCAACGCG--------GGTTGA----GAAAGCGGG-CGC---GACGC--T----GTGAAGTTT--TTCTTCCT-TGGATGTCTGATTCTTT---------
#=GS CryGCC/1-270 DE JALPCC010000061.1:30013-30282 Cryptococcus sp. GC_Crypt_2 iso00_70_61, whole genome shotgun sequence
CryGCC/1-270 ----------TCTAGATGATGAAAC-CTATTTCAAAACCATACGCAGTTTCCACTGAA-GCGTC--CTCCT--TTCGCC--GGGAG-TTTGT-ACCTATTCCGCATTTCCCCTTTTAGCTT--CCACAACAGAG-TA---GAGT-------GC------CCCTTTTGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCGATTGGG---AG------ATTC----CTCGACCGTGG--GCCTGT----------------------------------GAAA----------------------------------------GCAGGC--AGTCGGCAGAGAG-------GGTTGAGAA--GCGGTTTGG-GGC---GATGC---AATCTTGAAGTTTT-TTCTTCCT-TGGATGTCTGATTCTT----------
#=GS TakKor/1-284 DE BCKT01000013.1:865986-866269 Takashimella koratensis DNA, scaffold: scaffold_12, strain: JCM 12878, whole genome shotgun sequence
TakKor/1-284 --------CTTCAAGATGAAACCAA-GTTT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCC---CTGCCA-GGGAG-TTTGTTCCCCATCGTACCCGAAACCTCCCCTTTCCTCCGCGACCGGGTCGCA-GTG------CACCTT-----CTTTTGTCTC--------CGATATGAACCTATTG-----TT---GAGACGGGGG-----T-------CGC--AACTCCCCAACTG--TCGG------------------------------GCTC--GTGA---GAGC------AG---------------------------TCGG--AAGGGTGGATGG---------GATTAA----GGCGGATGG-GGC---GATGC--T-AGTTTGAAGTTT--TTCTTCCT-TGGATGTCTGATCTGTTCTCTT----
#=GS Amyc-1/1-257 DE CP053623.1:854775-855031 Apiotrichum mycotoxinovorans strain CICC 1454 chromosome 4
Amyc-1/1-257 ------GCTATCATGATGAAACCAC-TTGT---AAAACCATACGCAGTTTCCACTGCA-GCGTC--CTCC----CTGCCA--GGAG-TTTGTT-T-CCGCGCCGCGCACCGCCGAGCCG--------CCGATGG-TT---GAGTGC----------------CCCGGTTC---------CGGTTGAACCTCCG------TT---GAGCCGGG------C-----GCATTC----CCTGAGG------GCCTTC----------------------------CTCT---TC----GGAG------TC-------------------------GAGGGC--AGCTGGCAGACG---------CGATTG-----CAACGCCG-GGC---GATGC--T-A---TGAAGTTT-TCTCTTCCT-TGGATGTCTGAATCAACTTGTC----
#=GS DioCro/1-248 DE BCKK01000002.1:1565420-1565667 Dioszegia crocea DNA, scaffold: scaffold_1, strain: JCM 2961, whole genome shotgun sequence
DioCro/1-248 --------TTCTATGATGATATACC-ATAC---AAAACCATACGCAGTTTCCACTGAA-GCGTC--CTCCT--TTCGCC--GGGAG-TTTGT-CCCTGTCCTTTGTGGATCCCAGATC-------TCCCCCGGG-TC---GAGT-------C--------GTGTCAGCTC---------CGGATGAACCTTCTG-----TT---GAGCTGGCAT-----TG-----ATTC----TCTGGGTGG----CCTCT------------------------------------CC------------------------------------------GGAGG--AGCCGGTCTGTG---------GTGACG----CGGGGACGG-GGC---GATGC---ATACTTGAAGTTT--TTCTTCCT-TGGATGTCTGACTCT-----------
#=GS DioHun/1-246 DE JAKWFO010000005.1:c941888-941643 Dioszegia hungarica strain PDD-24b-2 MKK02contig_8, whole genome shotgun sequence
DioHun/1-246 --------TTCTATGATGAAACCCA-TTAC---AAAACCATACGCAGTTTCCACTGAA-GCGTC--CTCCT--TTCGCC--GGGAG-TTTGT-CCCTGTCTTTTCCAGATCCCAGACCC-------CCTCCGAG-TC---GAGT-------C--------ACGCCAGTTC---------CGAATGAACCTTCG------TT---GAGCTGGCGT-----CG-----ATTC----CTCGGGGG-----CCTCT------------------------------------TC------------------------------------------GGAGG--AGCAGGTCTGTG---------GGGATG----GGGAGACGG-GGC---GATGC---ATACTTGAAGTTT--TTCTTCCT-TGGATGTCTGACTCT-----------
#=GS KwoSha/1-293 DE NQVO01000010.1:c537604-537312 Kwoniella shandongensis strain CBS 12478 scaffold00010, whole genome shotgun sequence
KwoSha/1-293 ------TTTACAATGATGAAATTTT-CTAT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCT--TTCGCC--GGGAG-TTTGT-CCTCGTTCCTCTCTCCCTCCCTCTACTT--CCACACCGGAG-TT---GAGTCTG---CCTT-----------CGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCG--------TCGG--TAGATTC----CTCGACCGTGG--GCTCAGGTC----------------AGAAAA---CCTT---TC----GAGG---CGATTAC--------------------GGCCTGAGC--AGTCGGTGAGGTT--------GTGGAG----AGTTGACGA-GGC---GATGC--T-AGTTTGAAGTATT-TTCTTCCT-TGGATGTCTGACTCTTT---------
#=GS KocImp/1-274 DE NBSH01000014.1:c376511-376238 Kockovaella imperatae strain NRRL Y-17943 BD324scaffold_14, whole genome shotgun sequence
KocImp/1-274 ---------TTCATGATGAAACATC--TGTC--AAAACCATACGCAGTTTCCACTGAATGCGTC--CTCCT---TTACTC-GGGAG-TTTGTGTTCTCTCTCTGAAGCCTCGAGCCT---------CTCGTGGG-TT---GAGCTA--------------CCTGGTTCCC---TC------GTCGAACCTAC-------TT---GGGT-CCGGG----------TGGTTC----CCCATGTG-----GCGACG------------TTC-----GAAAC---CTC---TCAC----GAG---CTATTA---GAG--------T---------CGTTGC--AGCCGGATCGAGT--------GAGGAA----AGGATTGA--GCC---GATGC---GAAATTGAAGCTT-TTTCTTCCT-TGGATGTCTGATCCCT----------
#=GS TreMes/42-328 DE Treme1:TREMEscaffold_7:1173203:1173821:-1
TreMes/42-328 ------GTCGAAATGATGAAACATA-TTTT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCC---CTGCCA-GGGAG-TTTGTA--CCCGTTCCTCCACCCCCGTTCTCCTC-----CAC------AACC-GAG--------TC--GGACTTC-TCCATTC--------CGGGTTGAACCTACCTG----TT---GAGTGTTGAAGTCC---------CTC-AACC----GTG-----GGCGCGGTCC----------------GAAA--ACC------TA-------GGT--GATTA--------------------GGATTGTGTC--AGCCGGTAAACGG--------GACGAG----GAGGGATGG-GGC---GATGC--T-ATTTTGAAGTTT--CACTTCCT-TGGATGTCTGATCTACTCGTCT----
#=GS NaeAur/1-284 DE JAKFAO010000011.1:939834-940117 Naematelia aurantialba strain NX-20 Contig11, whole genome shotgun sequence
NaeAur/1-284 ----TCTCCTACAAGATGAAACATC--TGTC--AAAACCATACGCAGTTTCCACTGAATGCGTC--CTCTC--TTCG----GGGAG-TTTGTGCCTGTCCCGGCATCTCTTCGCACTT--------CCATCGAG-TC---GAGTTTC---------------CCTCGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCGGGG------T----GAGATTC----CTCGTTGG-----GCCCACAGCACC--------------GAAA--GCC------TC-------GGC--GACTA----------------GGAGCTGTTGTGGC--AGTCGGTGCGGC----------AAGGT----GACGAGCGG-GCC---GATGC--T-AAATTGAAGTTT-TTTCTTCCT-TGGATGTCTGAACCATT---------
#=GS TreFuc/1-291 DE BRDD01000039.1:c102650-102360 Tremella fuciformis NBRC 9317 DNA, KCNB80TF.39, whole genome shotgun sequence
TreFuc/1-291 ---TCCCTTTTCCTGATGAAATCCA-ATAC---AAAACCATACGCAGTTTCCACTGAATGCGTC--CTCCT--TTCGCC--GGGAG-TTTGCG-CTCCGTTCGTCCACCCTCTTTGGCCT------CTGCG----ACC--GGG--------TC---GAGCCCCCTTACTC--------GCGGTCGAACCTACTGT-----C---GAGTCTGGGAGTTC---------CCC-AAC---TGCAG-----GCGAC-CGTCT---------------GAAA--GCC-----CTT-------GGC--GATTA-------------------AGACGTGTCGC--AGCCGGTAAAGGA-------GGTTTGTG---GACGACAGA-GCC---GATGC--T-ACTTTGAAGTCTT-TTCTTCCT-TGGATGTCTGATTCTCC---------
#=GS TreYok/1-283 DE BRDC01000025.1:771240-771522 Tremella yokohamensis NBRC 100148 DNA, KCNB35TY.25, whole genome shotgun sequence
TreYok/1-283 ---------TTCCTGATGAAATCCA-ATAC---AAAACCATACGCAGTTTCCACTGAATGCGTC--CTCCT--TTCGCC--GGGAG-TTTGTG-CTCCGTTCGTCCACCCTCTTTGGCCT------CTGCG----ACC--GGG--------TC---GAGCCCCCTTACTC--------GCGGTCGAACCTACTGT-----C---GAGTCTGGGAGTTC---------CCC-AAC---TGCAG-----GCGAC-CGTCT---------------GAAA--GCT-----TTT-------GGC--GATTA-------------------AGACGTGTCGC--AGCCGGTAAAGGA-------GGTTTGTG---GACGGCAGA-GCC---GATGC--T-AGTTTGAAGTCTT-TTCTTCCT-TGGATGTCTGATTCT-----------
#=GS SaiPod/1-278 DE CABWHZ010000056.1:75898-76175 Saitozyma podzolica genome assembly, contig: NODE_56_length_182117_cov_29.0947, whole genome shotgun sequence
SaiPod/1-278 ----------TAATGATGAAATCCA-TTTC---AAAACCATACGCAGTTTCCACTGAA-GCGTC--CTCCT--TTCGCC--GGGAG-TTTGTA-CCCATCGTTTCGTGCCCCTCTTCTTCCT----CTACCGG--TC---GAGC-------AA--------GCCGGGTTC---------CGTGTGAACCTCACG-----TC---GAGCCCGGT------GA-----GTTC-----CCGGTGG-----GCGAGGTACC----------------GAAA--GCTCCT--TTGC---GGGGGT--GATTA--------------------GGT-CCTCGC--AGTCGGGAGAGT--------GGT-GGT----GCGCGGTGG-GGC---GATGC--T-AAATTGAAGTATT-TTCTTCCT-TGGATGTCTGAAATC-----------
#=GS SaiJCM/1-287 DE BCLC01000008.1:c335332-335046 Saitozyma sp. JCM 24511 DNA, scaffold_7, whole genome shotgun sequence
SaiJCM/1-287 --------TATTATGATGAAATCCA-ATTC---AAAACCATACGCAGTTTCCACTGAA-GCGTC--CTCCT--TTCGCC--GGGAG-TTTGTA-CCCACCGTTTCGTGCCCCTCTTCTTCCT----CTGCCGG--TC---GAGC-------AA--------GCCGGGTTC---------CGTGTGAACCTCACG-----TC---GAGCCCGGC------GA-----GTTC-----CCGGTGG-----GCGAGGTATC----------------GAAA--GCTCC--TTTGCC---GGGGT--GACTA--------------------GAT-CCTCGC--AGTCGGGACGGT--------GGT-GGT----GCGCGGTGG-GGC---GATGC--T-AAATTGAAGTATT-TTCTTCCT-TGGATGTCTGAATCACACAGCT----
#=GS PapTer/1-268 DE JAHXHD010001261.1:47481-47748 Papiliotrema terrestris strain LS28 scaffold-1260, whole genome shotgun sequence
PapTer/1-268 ------TCTCGAATGATGAAAACAT-CTGT---AAAACCATACGCAGTTTCCACCGAATGTGTC--CTCCCC--TACTC-GGGGAG-TTTGTA-CCCATTCTCACCACCTTTCCTCCACT------CTGCCGGG-TCGC-AGCCCT----CTGT-----------CGT-----AA----GAGTTGAACCTCTC------GA---GCG-----------CTT---AGGGTT--C-CCCGGCGG-----GC-------------------------TT-----TGGG--CTCGT--CTCA------TT-----------------------------GC--AGTCGTGGGGTC--------GAGAGGAG---GTGGTTTGG-GGC---GATGC---GAATTTGAGGTTT-ATTCTTCCT-TGGATGTCTGACTCTTCTTTT-----
#=GS CryFla/1-260 DE CAUG01000622.1:c82520-82261 Cryptococcus flavescens NRRL Y-50378 WGS project CAUG00000000 data, contig NODE_1791_length_325247_cov_46_176311, whole genome shotgun sequence
CryFla/1-260 ------TCTTGAATGATGAAACATC--TGT---AAAACCATACGCAGTTTCCACCGAATGTGTC--CTCCCC--TACTC-GGGGAG-TTTGTA-CCCTCTCTCACT-CCTTTCCTCCGCT------CTGCCGGG-TCG--AGCCCT----CTGC-----------CGC-----AA----GAGTTGAACCTCTC------GA---GCG-----------CTT---AGGGTT--C-CTCGGTGG-----GC-------------------------TT-----TGAG--CTCGT--CTCA------TC-----------------------------GC--AGCTGTGGGGTT--------GAGAGGCG---GTGGCTTGG-GGC---GATGC---GAATTTGAGGTTT-ATTCTTCCT-TGGATGTCTGACTCTT----------
#=GS PapLau/1-327 DE JDSR01000088.1:c42103-41777 Papiliotrema laurentii RY1 contig_93, whole genome shotgun sequence
PapLau/1-327 --------TTGAATGATGAAATGTT-TTTT---AAAACCATACGCAGTTTCCACTGAATGCGTC--CTCCCT--TACTC-GGGGAG-TTTGTA-CCCATTCTC-CGGACCTCCTCAACCT------CCTCGAAG-TCGC-AGCCCC-----CC-------GTT-TTGTGA---------CAGTTGAACCTCTCG------A---TCA-GGCGGC----------GGGGTT--C-CTTCTTGG-----GCTTGAGCTCTTTCCTCTCCC-----GAAC--GGCCTGC-TCAC--GCGGGTT-CTACTA---GGGGTCTCAAGAGGAAGAGTCACGAGC--AGTCGGTAGAGT--------CGAGAGAA---GGAGAGTGG-GGC---GATGC---GAATTTGAAGTTT-ATTCTTCCT-TGGATGTCTGACCCACCCTTCTT---
#=GS TreFJI/1-282 DE JAKLMH010000014.1:c20478-20197 Tremellales sp. FJII-L8-SW-PAB2 NODE_14_length_504163_cov_88.310461, whole genome shotgun sequence
TreFJI/1-282 -----TTTCGATATGATGAAACCTA-CTTC---AAAACCATACGCAGTTTCCACTGAA-GCGTC--CTCCT--TTCGCC--GGGAG-TTTGT-ACCCATCCCACCTTTCCCTTTATTGCTT--CCACAACAGAG-TCGT-AGCCCCC--TTGTTT---------CCATTC--------CGAGTTGAACCTCTCG-----TT---GAGTGG--------CTT--GGGAGTT--C-CTCGCCCGTGG--GCCGT-----------------------------TGGA--TCCG---TCCA------T---------------------------ATGGC--AGTCGGCGAAATT-------GGTCGAGAG--GAGGTTTGG-GGC---GATGC---AATCTTGAAGTTTC-TTCTTCCT-TGGATGTCTGATCAT-----------
#=GS KwoHea/1-305 DE ASQB01000005.1:221863-222167 Kwoniella heveanensis BCC8398 cont1.5, whole genome shotgun sequence
KwoHea/1-305 --------TCTTATGATGAAATATT-TGAT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCTT--TCGC--GGGGAG-TTTGT-CCTTGTTCTCTCTTGGTCCTCATC-ACTT-CCGCAACCGAG-TCGC-AGTCTTT---CTC------------CGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCG---------TTG--GGAGATT--C-CTCAACCGTGG--GCTTG-TT------GAGCCT------GAAA----TATC--CTCGT--GATA----GATTA----GGGTTC---------------CGAGC--AGTCGGTGGTG---------TTGTCCG----AGGAGACTC-GGC---GATGC--T-AATTTGAAGTTTTTATCTTCCT-TGGATGTCTGATTCTTTCCTTCTTT-
#=GS FusOxy/1-292 DE WESS01000060.1:25637-25928 Fusarium oxysporum strain EtdFoc-17 scaffold87.1:size85144, whole genome shotgun sequence
FusOxy/1-292 --------TTGTAGGATGAAACATC-TGT----AAAACCATACGCAGTTTCCACTGAATGTGTC--CTCCC---CTGCCA-GGGAG-TTTGTA-CCCATCCTCGTCTCCTTTCCTCCA-T------CCTTCGGG-TTGT-AGCC-------CT---------CTGCGT-----AA----GAGTTGAACCTCTC------GA---GCGCAG---------TT-----GGTT--C-CTCGTTGG-----GCTTTTTCTCC---------------GAAC--GCTTCT-TCCTTC--GGGGGT-TGTCTACTAG---------------GGCGGGACTGC--AGCCGTGGGGTTGA-------GAGGAG----GTGGTTTGG-GGC---GATGC---GAATTTGAAGTCT-ATTCTTCCT-TGGATGTCTGACCTTTAT--------
#=GS KwoMan/1-320 DE ASQF01000012.1:416555-416874 Kwoniella mangroviensis CBS 8886 cont1.12, whole genome shotgun sequence
KwoMan/1-320 ---------TCTATGATGAAATGTT-TCTT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCTT--TCGC--GGGGAG-TTTGTTCCTTCTTTCTTTTCAGTCTTTCTCTACCT-TCATAACTGAG-TTGC-AGTTCCT---CTT------------CGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCG--------TCGTT-GGGAATT--C-CTCGACTTTGG--GCTCACCCCGAAGTTCTCATT----CGAAAA---TCCT---TC----GGGA----CGACTA-----GATTGTCGATTGAAGGGACCGAGC--AGTCGGTAGATTG--------AGATTG----GAGAAATTA-GGC---GATGC--T-AAATTGAAGTTTTTTTCTTCCT-TGGATGTCTGATTCT-----------
#=GS KwoDej/1-317 DE ASCJ01000008.1:601320-601636 Kwoniella dejecticola CBS 10117 cont1.8, whole genome shotgun sequence
KwoDej/1-317 --------TTCAATGATGAAATGTT-TTTT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCTT--TCGC--GGGGAG-TTTGT-CCTTCTTCCCCTTGATCTCTTTCT-ACTA-CCACGACTGAG-TTGC-AGTTCCC---CTT------------CGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCG---------TCG--GGGAATT--C-CTCGACTGTGG--GCTCGATCATACATGACTCAAGCC--GAAC---GTCTC---TC----GGGAT---TATTA-------GGTTTGCAGTCTTTGGTCGTTGC--AGTCGGTAGACC--------GAGATCG----GGGGAAGTA-GGC---GATGC--T-AATTTGAAGTTTT-TTCTTCCT-TGGATGTCTGATTCT-----------
#=GS KwoPin/1-317 DE ASCL01000030.1:c95867-95551 Kwoniella pini CBS 10737 cont1.30, whole genome shotgun sequence
KwoPin/1-317 --------TTGAATGATGAAATGTT-TTTT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCTT--TCGC--GGGGAG-TTTGTTCCTTTTTCTTCCTGATCTTCTTTC-ACTT-CCATAACTGAG-TTGT-AGTTCCC---CTC------------CGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCG---------TTT--GGGAATT--C-CTCGAATTTGG--GCTCGACCATATATGACCCAGATC--GAAC---ATCTT---TC----GGGAT---TACTA-------GATTTGCGGTT-TTGGTTGTTGC--AGTCGGTAGATC--------AAGATTG----GGGGAATTA----GGCGATGCT---AATTTGAAGTTTT-TTCTTCCT-TGGATGTCTGATTCT-----------
#=GS KwoBes/1-317 DE ASCK01000013.1:1006639-1006955 Kwoniella bestiolae CBS 10118 cont1.13, whole genome shotgun sequence
KwoBes/1-317 ---------TCTATGATGAAATGTT-TTTT---AAAACCATACGCAGTTTCCACTGAACGCGTC--CTCCTT--TCGC--GGGGAG-TTTGT-CCTTCTTCCTTCCATCTCCTTCT--ACTT-CCATAACTGAG-TTGT-AGTTCCT---CTT------------CGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCG--------TTGT--GGGAATT--C-CTCGACCTTGG--GCTCGCTCATATACACCATATTTC--GAACA---TCCT---TC----GGGA----CTACTA------GAA-TGTTGGTCTTGGGTTGAGC--AGTCGGTAGATTG--------AGATTG----GAGGGATTA----GGCGATGCT---AAATTGAAGTTTT-TTCTTCCT-TGGATGTCTGATTCTTT---------
#=GS CryDep/76-328 DE Filo_depa_CBS7841_V1:supercont1.34:136223:136840:-1
CryDep/76-328 ------ACCTATCTGATGAAATGTT-TATTC--AAAACCATACGCAGTTTCCACCGATCGCGTC--CTCC----CTGCCA--GGAG-TCTGT-CTT-GGCTTTGCTGCTTCAATTTACCT--------CTAGGTCAGAC-TATCA------TT------------AGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCT----------TT----TGATG-----------------CTGTTTGA------------------CTT------GGG---CT----CTT--------------------------------TTGAGCAG----ACGGTGTTTTGTTGCACAAAGCT------------------TAGCGATGCA---AA---GAGGTCTT-TACTTCCT-TGGATGTCTGATTATCTTTTTT----
#=GS CryDep-2/1-254 DE AWGL01000010.1:c143891-143638 Cryptococcus depauperatus CBS 7855 supercont2.10, whole genome shotgun sequence
CryDep-2/1-254 ---------TATCTGATGAAATGTT-TATTC--AAAACCATACGCAGTTTCCACCGATCGCGTC--CTCC----CTGCCA--GGAG-TCTGT-CTT-GGCTTTGCTGCTTCAATTTACCTCT--------AGGTCAGAC-TATCA------TT------------AGTTC--------CGAGTTGAACCTCTCG-----TT---GAGCT----------TT----TGATG---------A--------TGTCTGA------------------CTT------GGG---CTC---TTT---------------------------------TGAGCA----GACGGTGTTTTGTTGCACAAAGCT------------------TGGCGATGCA---AA---GAGGTCTT-TACTTCCT-TGGATGTCTGATTACCTTTTTTTTCT
Annotation/1-121 ------------RUGAUGA------------ -GUAAGCGUCAAAG--CUGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------RUGAUGA-------------------GGAAGGA-ACCUACACUGA---------------
Hsap-a/1-86 ---------ACTGTGATGATGG---TTTTCCA-----ACATTCGCAGTTTCCACCAG-------------------------------------------------------------------------------------------------------------------------------AAAGGTTTTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CT-TAGTGTTGGGT-------AAA-------CCTTCCT-TGGATGTCTGAGT-------------
Hsap-b/1-87 ---------ACTATGATGATTGG---TTGCCAG----ACATTCGCAGTTTCCACCAG-------------------------------------------------------------------------------------------------------------------------------AAATGTTTTTC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CT-T-ATGTTGGCC--------AG------TTCTTCCT-TGGATGTCTGAGT-------------
#=GC SS_cons -----------------------------------------------------------(((((--(((((----------)))))------------------------------------((((---(((((------(((((--------------((((--(((((--------((((--------))))----------)))))--))))----------)))))----)))))--))))--((((((----------------------------((((---------))))---------------------------------))))))-------------------------------------------------)))))-----------------------------------------
//
Image source snR190:
# STOCKHOLM 1.0
#=GF RW van Nues, May 2023
#=GF https://coalispr.codeberg.page/paper
#=GS JEC21-snR190/1-144 DE AE017344.1:194962-195105 Cryptococcus neoformans var. neoformans JEC21 chromosome 4 sequence
JEC21-snR190/1-144 --------------TCGAAGTGATGACCTTC------ATTCTCTCCCCGTCTAATCGACCATGGCGA-ATGAGTTAGGGCG-CTCCTTCGCAGGAGCTGCCTGGAACCCCTTCCTC----------TCTTTT--------GAGGACGGGCTTATGGCGCTTGCCAGCGCTTT--TGTCTGATCTCC---
#=GS H99-mRNA/1-142 DE GJKG01007043.1:88-229 TSA: Cryptococcus neoformans var. grubii isolate H99 CNAG_00930, transcribed RNA sequence
H99-mRNA/1-142 --------------TCGAAGTGATGACCTTC------ATTCTCTCCCCGTCTAATCGACCATGGCGA-ATGAGTTAGGGCG-CTCCTTCGCAGGAGCTGCCTGGAACCCCTTCCTC-----------TTTT---------GAGGACGGGCTTATGGCGCTTGCCAGCGCTTT--TGTCTGATCTCC---
#=GS EN28/1-142 DE CP025721.1:c1592771-1592630 Cryptococcus neoformans strain EN28 chromosome 5, complete sequence
EN28/1-142 --------------TCGAAGTGATGACCTTC------ATTCTCTCCCCGTCTAATCGACCATGGCGA-ATGAGTTAGGGCG-CTCCTTCGCAGGAGCTGCCTGGAACCCCTTCCTC-----------TTTT---------AAGGACGGGCTTATGGCGCTTGCCAGCGCTTT--TGTCTGATCTCC---
#=GS R265/1-143 DE CP025772.1:174034-174176 Cryptococcus gattii VGII R265 chromosome 14, complete sequence
R265/1-143 --------------TCGATGTGATGACCTTC------ATTCTCTCCCCGTCTAACCGACCATGGCGA-ATGAGTTAGGGCG-CTCCTTCGCAGGAGCTGCCTGGAACCCCTTCCTC-----------TTTTT--------GAGGACGGGCTTATGGCGCTTGCCAGCGCTTT--TGTCTGATCTCC---
#=GS WM276/1-142 DE CP000295.1:c322619-322478 Cryptococcus gattii WM276 chromosome J, complete sequence
WM276/1-142 --------------TCGATGTGATGACCTTC------ATTCTCTCCCCGTCTAACCGACCATGGCGA-ATGAGTTAGGGCG-CTCCTTCGCAGGAGCTGCCTGGAACCTCTTCCTC-----------TTTT---------GAGGACGGGCTTATGGCGCTTGCCAGCGCTTT--TGTCTGATCTCC---
#=GS CryWin/1-140 DE CP034264.1:1386327-1386466 Cryptococcus wingfieldii strain CBS7118 chromosome 4, complete sequence
CryWin/1-140 --------------TCGACGTGATGACCTTC------ATTCTCTCCCCGTCTAATCGACCATGGCGA-ATGAGTTAGAGCG-CTCCTTCGCAGGAGATGCTTGGAACCCCTCCCTC------------TC----------GAGGGCGGGTTCATGGCGCTTGCCAGTGCCTT--TGTCTGATCTCC---
#=GS CryAmy-1/1-140 DE XM_019136337.1:68-207 Cryptococcus amylolentus CBS 6039 argininosuccinate synthase, variant 1 (L202_02683), mRNA
CryAmy-1/1-140 --------------TCGACGTGATGACCTTC------ATTCTCTCCCCGTCTAATCGACCATGGCGA-ATGAGTTAGAGCG-CTCCTTCGCAGGAGATGCTCGGAACCCCTCCCTC------------TC----------GAGGGCGGGTTCATGGCGCTTGCCAGTGCCTT--TGTCTGATCTCC---
#=GS CryAmy-1_mRNA/1-140 DE XM_019136337.1:68-207 Cryptococcus amylolentus CBS 6039 argininosuccinate synthase, variant 1 (L202_02683), mRNA
CryAmy-1_mRNA/1-140 --------------TCGACGTGATGACCTTC------ATTCTCTCCCCGTCTAATCGACCATGGCGA-ATGAGTTAGAGCG-CTCCTTCGCAGGAGATGCTCGGAACCCCTCCCTC------------TC----------GAGGGCGGGTTCATGGCGCTTGCCAGTGCCTT--TGTCTGATCTCC---
#=GS CryAmy_mRNA/1-140 DE XM_019136336.1:68-207 Cryptococcus amylolentus CBS 6039 argininosuccinate synthase (L202_02683), mRNA
CryAmy_mRNA/1-140 --------------TCGACGTGATGACCTTC------ATTCTCTCCCCGTCTAATCGACCATGGCGA-ATGAGTTAGAGCG-CTCCTTCGCAGGAGATGCTCGGAACCCCTCCCTC------------TC----------GAGGGCGGGTTCATGGCGCTTGCCAGTGCCTT--TGTCTGATCTCC---
#=GS ApiMyc/1-143 DE CP053623.1:854478-854620 Apiotrichum mycotoxinovorans strain CICC 1454 chromosome 4
ApiMyc/1-143 --------------TCAACCTGATGACCTTC-----TATTCTCTCCCCGTCTTACCGACCATGTCGA-ACGAGATAGGGAC-CCCCTTCGCGGGGGGCTCCTGGACCCCTTCGCCTC----------GCAA---------GAGGCTGGGCTAATGGCGCTTGCCAGTGCCTT--CGTCTGATCCT----
#=GS ApiMyc-b/1-143 DE CP049826.1:c2195733-2195591 Apiotrichum mycotoxinovorans strain GMU1709 chromosome VI
ApiMyc-b/1-143 --------------TCAACCTGATGACCTTC-----TATTCTCTCCCCGTCTTACCGACCATGTCGA-ACGAGATAGGGAC-CCCCTTCGCGGGGGGCTCCCGGACCCCTTCGCCTC----------GCAA---------GAGGCTGGGCTAATGGCGCTTGCCAGTGCCTT--CGTCTGATCCT----
#=GS VanPse/1-143 DE CP086717.1:c2552692-2552550 Vanrija pseudolonga isolate DUCC4014 chromosome 4
VanPse/1-143 --------------TTGATCTGATGACCTTC----TTATTCTCTCCCCGTCTTACCGACCATGTCGA-ACGAGATAGGGAC-CCCCTTCGCGGGGGGCTCCCGGACCCCTTCGTGC------------TT----------GCACGCGGGCTAATGGCGCTTGCCAGTGCCTT--TGACTGATCCCCT--
#=GS PapLau/1-148 DE GFWE01018345.1:90-237 TSA: Papiliotrema laurentii RY1 YE1_c5499_g1_i1 transcribed RNA sequence
PapLau/1-148 --------------TCACCCTGATGACCTTC-----TATCCTCACCCCGTCTTATCGACCATGTCGA-ACGAGTTAGGCCTCCCCCTTCGCGGGGGGAGGCTGGAACCCCTTTCTTCC----------TC--------GGAAGAACGGGCTTAT-GCGCTTGCCAGTGCTTT--TGTCTGATCCACCCT
#=GS PapLau-2nd/1-143 DE GFWE01018345.1:230-372 TSA: Papiliotrema laurentii RY1 YE1_c5499_g1_i1 transcribed RNA sequence
PapLau-2nd/1-143 -------------TCCACCCTGATGACCTTC-----TATCCTCACCCCGTCTTATCGACCATGTCGA-ACGAGTTAGGCCTCCCCCTTCGCGGGGGGAGGCTGGAACCCCTTTCTTCC----------TC--------GGAAGAACGGGCTTAT-GCGCTTGCCAGTGCTTT--TGTCTGATC------
#=GS PapLau-b/1-144 DE GFWE01000923.1:90-233 TSA: Papiliotrema laurentii RY1 AS1_c1114_g1_i1 transcribed RNA sequence
PapLau-b/1-144 --------------TCACCCTGATGACCTTC-----TATCCTCACCCCGTCTTATCGACCATGTCGA-ACGAGTTAGGCCTCCCCCTTCGCGGGGGGAGGCTGGAACCCCTTTCTTCC----------TC--------GGAAGAACGGGCTTAT-GCGCTTGCCAGTGCTTT--TGTCTGATCCT----
#=GS PapTer/1-146 DE JAHXHD010001261.1:47189-47334 Papiliotrema terrestris strain LS28 scaffold-1260, whole genome shotgun sequence
PapTer/1-146 ----------------TCAGTGATGACCTTC-----TATCCTCACCCCGTCTTACCGACCATGTCGA-ATGAGTTAGGCCTCTCCCTTCGCGGGGAGAGGCTGGACCCCCTCCCTCAT---------TTGC-------GTGAGGGCGGGCTCAT-GCGCTTGCCAGCGCTTT--TGTCTGATCCATT--
#=GS ApiBra/1-146 DE JAMALJ010000038.1:c152040-151894 Apiotrichum brassicae strain M2204 ctg_38, whole genome shotgun sequence
ApiBra/1-146 ------------TCCCAACCTGATGACCTTC----TTATTCTCTCCCCGTCTTATCGACCATGACGA-ACGAGATAGGGA-CCCCCTTCGCGGGGGGCTCCCGGACCCCCGTG--------------CCG---------CACGGCCGGGCTAATGGCGCTTGCCAGTGCCTT--TGTCTGATCCACAAC
#=GS ApiSia/1-137 DE JALJEG010000016.1:c157983-157847 Apiotrichum siamense strain L8in5 NODE_16_length_432637_cov_11.418381, whole genome shotgun sequence
ApiSia/1-137 -----------------TCCTGATGACCTTC-----TATTCTCTCCCCGTCTTACCGACCATGTCGA-ACGAGATAGGGA-CCCCCTTCGCGGGGGGCTCCCGGACCCCTGTGC-------------TCT------------GCACGGGCTCGCGGCGCTTGCCAGTGCCTT--TGTCTGATCCCACT-
#=GS VanHum/1-143 DE QKWK01000005.1:904414-904556 Vanrija humicola strain CBS 4282 CBS4282_scaffold05, whole genome shotgun sequence
VanHum/1-143 --------------TTGATCTGATGACCTTC----TTATTCTCTCCCCGTCTTACCGACCATGTCGA-ACGAGATAGGGA-CCCCCTTCGCGGGGGGCTCCCGGACCCCTTCGTGC------------TT----------GCACGCGGGCTAATGGCGCTTGCCAGTGCCTT--TGACTGATCCCCT--
#=GS VanFra/1-146 DE BEDY01000003.1:c403442-403297 Vanrija fragicola DNA, scaffold: scaffold_3, strain: JCM 1530, whole genome shotgun sequence
VanFra/1-146 -------------TCTACTCTGATGAACTTC----TTATTCTCTCCCCGTCTAACCGACCATGTCGA-ATGAGATAGGAA-CCCCCTTCGCGGGGGGCTTCCGGACCCCCTCCTGTGC----------TT--------GCACAGGCGGGCTTATGGCGCTTGCCAGTGCCTT--TGTCTGATCCT----
#=GS ApiPor/1-154 DE RSCE01000002.1:831457-831610 Apiotrichum porosum strain DSM 27194 scaffold_2, whole genome shotgun sequence
ApiPor/1-154 ---------------TGATCTGATGACCTTC-----TATTCTCTCCCCGTCTTACCGACCATGTCGA-ACGAGATAGGAA-CCCCCTTCGCGGGGGGCTTCCGGACCCCGGCCACTTGCAC------TTTT----GTGCTTGGGGCGGGCTTACGGCGCTTGCCAGTGCCTT--TGTCTGATCCCACC-
#=GS TriAsa/1-141 DE AMBO01000411.1:24255-24395 Trichosporon asahii var. asahii CBS 8904 contig0412, whole genome shotgun sequence
TriAsa/1-141 --------------TCGAAATGATGACCTTC----TATTTCTCTCCCCGTCTTATCGACCATGACGA-ACGAGACAGG-AGCCCCCTTCGCAGGGGGCT-CTGGACCCCCTCGCGGC-----------TT----------GCCGCAGGGCTAATGGCGCTTGCCAGCGCTTA--AGTCTGATCCC----
#=GS NaeEnc/1-149 DE MCFC01000036.1:c69967-69819 Naematelia encephala strain 68-887.2 BCR39scaffold_36, whole genome shotgun sequence
NaeEnc/1-149 --------------TCGAAGTGATGACCTTC------ATTCTCTCCCCGTCTAACCGACCATGGCGA-ACGAGCTAGGGATCCCCCTTCGCGGGGGGATTCTGGACCCCCTTCCTCTGCT--------TT------GGTAGAGGACGGGTTTAT-GCGCTTGCCAGCGCTTT--TGTCTGATCACCT--
#=GS NaeAur/1-149 DE JAKFAO010000011.1:939563-939711 Naematelia aurantialba strain NX-20 Contig11, whole genome shotgun sequence
NaeAur/1-149 --------------TCGAAGTGATGACCTTC------ATTCTCTCCCCGTCTAACCGACCATGGCGA-ACGAGCTAGGGATCCCCCTTCGCGGGGGGATCCCGGACCCCCTTCCTCTGCT--------TT------AGCAGCGGACGGGCTTAT-GCGCTTGCCAGCGCTTT--TGTCTGATCACCT--
#=GS CutOle/1-136 DE QKWL01000003.1:455356-455491 Cutaneotrichosporon oleaginosum strain ATCC 20508 ATCC20508_contig03, whole genome shotgun sequence
CutOle/1-136 --------------TCACCCGGATGACCTTC-----TATTCTCTCCCCGTCTTACCGACCATGTCGA-ACGAGATAGGG-CTCCCCTTCGCGGGGGAGTC-TGGAACCCCGCTA--------------C-------------ATGCGGGCTCTCGGCGCTTGCCAGTGCCTT--TGTCTGATCCCTC--
#=GS SaiPod/1-145 DE CABVTU010000500.1:c20511-20367 Saitozyma podzolica genome assembly, contig: NODE_503_length_34331_cov_8.25722, whole genome shotgun sequence
SaiPod/1-145 --------------TTGACAAGATGACCTTC-----TATTCTCTCCCCGTCTAATCGACCATGGCGA-ACGAGATAGGG-CGCTCCTTCGCGGGAGTCGTCTGGAACCCCTCCTCGGAC--------CTC-------GTCCGGGGCGGGCTCAT-GCGCTTGCCAGCGCTTT--AGTCTGATCC-----
#=GS TakaKor/1-140 DE BCKT01000013.1:865655-865794 Takashimella koratensis DNA, scaffold: scaffold_12, strain: JCM 12878, whole genome shotgun sequence
TakaKor/1-140 --------------TCGACCTGATGACCTTC------ATTCTCTCCCCGTCTTACCGACCATGTCGA-ACGAGACAGGG-GCCTCCTTCGCGGGAGTGCTCCGGATCCCCTCTGC-------------C-------GCAAGGCAGCGGGCTCTT-GCGCTTGCCAGCGCTTT--CGTCTGATCCC----
#=GS Ram5/1-143 DE ASCM01000098.1:c27362-27220 Cryptococcus gattii VGII Ram5 cont1.98, whole genome shotgun sequence
Ram5/1-143 ------------TCTCGATGTGATGACCTTC------ATTCTCTCCCCGTCTAACCGACCATGGCGA-ATGAGTTAGGGC-GCTCCTTCGCAGGAGCTGCCTGGAACCCCTTCCTCTTT---------TT----------GAGGACGGGCTTATGGCGCTTGCCAGCGCTTT--TGTCTGATCT-----
#=GS EJB2/1-142 DE ATAL01000133.1:c10327-10186 Cryptococcus gattii EJB2 cont1.133, whole genome shotgun sequence
EJB2/1-142 --------------TCGATGTGATGACCTTC------ATTCTCTCCCCGTCTAACCGACCATGGCGA-ATGAGTTAGGGC-GCTCCTTCGCAGGAGCTGCCTGGAACCTCTTCCTCTTT----------T----------GAGGACGGGCTTATGGCGCTTGCCAGCGCTTT--TGTCTGATCTCC---
#=GS CA1280/1-144 DE ASCN01000092.1:173918-174061 Cryptococcus gattii CA1280 cont1.92, whole genome shotgun sequence
CA1280/1-144 ------------TCTCGATGTGATGACCTTC------ATTCTCTCCCCGTCTAACCGACCATGGCGA-ATGAGTTAGGGC-GCTCCTTCGCAGGAGCTGCCTGGAACCTCTTCCTCTTT----------T----------GAGGACGGGCTTATGGCGCTTGCCAGCGCTTT--TGTCTGATCTCC---
#=GS IND107/1-143 DE ATAM01000083.1:139453-139595 Cryptococcus gattii VGIV IND107 cont2.83, whole genome shotgun sequence
IND107/1-143 --------------TCGATGTGATGACCTTC------ATTCTCTCCCCGTCTAACCGACCATGGCGA-ATGAGTTAGGGC-GCTCCTTCGCAGGAGCTGCCTGGAACCCCTTCCTC-----------TTTTT--------GAGGACGGGCTTATGGCGCTTGCCAGCGCCTT--TGTCTGATCTCC---
#=GS DioCro/1-153 DE BCKK01000002.1:1565136-1565288 Dioszegia crocea DNA, scaffold: scaffold_1, strain: JCM 2961, whole genome shotgun sequence
DioCro/1-153 -------------TCCGATAAGATGACCTTC-----TATTCTCTCCCCGTCTAACCGACCATGTCGA-ATGAG-CAGGG-CATGCCTTCGCGGGTGTG-CCCGGACCCCCAACCTCTGGAAGT----CTG----ACTTCCAGGGGAGGGCTTTT-GCGCTTGCCAGCGCTTT--TGTCTGATCTCC---
#=GS DioHun/1-159 DE JAKWFO010000005.1:c942192-942034 Dioszegia hungarica strain PDD-24b-2 MKK02contig_8, whole genome shotgun sequence
DioHun/1-159 ------------TACCGAAAAGATGACCTTC-----TATTCTCTCCCCGTCTAACCGACCATGTCGA-ATGAG-CAGGG-CATGCCTTCGCGGGTGTG-CCCGGAACCTCAACCCCTTTGATTCCC--CC-GGGAACCGAAGGGGAGGGCTTTT-GCGCTTGCCAGCGCTTT--TGTCTGATCCCT---
#=GS CryDep/1-142 DE AWGK01000034.1:c137089-136948 Cryptococcus depauperatus CBS 7841 supercont1.34, whole genome shotgun sequence
CryDep/1-142 --------------TCGATGTGATGACCTTC-----TATTCTCTCCCCGTCTAATCGACCATGGCGA-ATGAGTTAGGGC-GCTTCTTCGCAGAAGCTGTCCGTACCCCCTTCCTC------------TTT---------GAGGACGGGTTTGTGGCGCTTGCCAGCGCTTT--TGTCTGATCCCT---
#=GS CryFla/1-147 DE CAUG01000622.1:c82816-82670 Cryptococcus flavescens NRRL Y-50378 WGS project CAUG00000000 data, contig NODE_1791_length_325247_cov_46_176311, whole genome shotgun sequence
CryFla/1-147 ---------------TTCAGTGATGACCTTC-----TATCCTCACCCCGTCTTACCGACCATGTCGA-ACGAGTTAGGACTCTCCCTTCGCGGGGANNNNNNNNNNCCCCCTCCCACAC--------GCAA-------GTGTGGGCGGGCTTAT-GCGCTTGCCAGCGCTTT--TGTCTGATCCAT---
#=GS TreTag/1-157 DE CAJHEQ010000407.1:c2982-2826 MAG: Tremellales sp. Tagirdzhanova-0007 genome assembly, contig: TREM_407, whole genome shotgun sequence
TreTag/1-157 TCCACACGCGAAGACCGAAAAGATGACCTTC-----TATTCTCTCCCCGTCTGACCGACCATGTCGA-ACGAGCTAGGGAA-CCCCTTCGCGGGGGCTTCCCGGATCCCCTTCCAG-----------TTTCT--------CTGGACGGGCTCAT-GCGCTTGCCAGCGCTTT--TGTCTGATCTCT---
#=GS FusOxy/1-149 DE WESS01000060.1:25345-25493 Fusarium oxysporum strain EtdFoc-17 scaffold87.1:size85144, whole genome shotgun sequence
FusOxy/1-149 -----------TCCTCAATCTGATGACCTTC-----TAATCTCTCCCTATCCTACCGACCATGCCGA-ACGAGTTAGGGCCCTCCCTTCGCGGGGAGGGACCGGACCCCCAGCCTCATT---------TC-------GGTGAGGTCGGGCTTTT-GCGCTTGCCAGCGCCTT--TGTCTGATTCT----
#=GS PhaRho/1-150 DE LN483157.1:c1978735-1978586 Phaffia rhodozyma Xanthophyllomyces dendrorhous genome assembly Xden1, scaffold Scaffold_69
PhaRho/1-150 --------------TCAACGTGATGACTTCAATCTTTTTTCTCTCCCCGTCCTATCGACCATGCCGAGACGACAGAGTTGGAGGGGGTCGCACCTCTCCAT-GGCGGACGCA---------------GCGA------TGCGTGTGCCGAGTTTTGAGGCTTGCCAGCCTCATTTTGCCTGATTCCT---
//